1. INTRODUCTION
Osteoarthritis (OA) is common in musculoskeletal diseases. OA occurs more commonly in the elderly. Approximately 303 million people worldwide suffered from OA in 2017 [1]. By 2050, more than 20% of the world’s population will be over the age of 60, 15% of whom will have symptoms of OA and more than a third of these will be disabled [2]. OA is quite popular in Vietnam but there are no exact statistics. Vietnam’s population aging rate is currently the fastest in Asia. It is predicted that by 2050, Vietnamese people aged 60 and over will be doubled from 11.9 to 29 million, accounting for about a third of the total population. Therefore, Vietnam will witness an increase in OA in the following years because OA is proportional to population aging.
MicroRNA (miRNA) is an RNA molecule with about 20-25 nucleotides that is able to regulate gene expression. Binding of miRNAs to its specific binding site on the 3’UTR region of the mRNA leads to mRNA hydrolysis or the inhibition of translation [3, 4]. Aberrant expression of miRNAs is reported in many diseases such as cancer and metabolic disorders [5]. Abnormal expression of several OA-associated miRNAs has been identified such as miR-483, miR-22, miR-9, miR-98, miR-485-5p, miR-455, miR-210, miR-27b, miR-140, and miR-29 [6-14].
miR-144-3p has been shown to be associated with OA. Specifically, miR-144-3p increased in OA chondrocytes [15] and 5-fold in OA osteoblasts [8]. Moreover, miR-144-3p inhibits CRAT which inhibits osteoclastogenesis [16]. Several genes involved in OA have also been shown to be the targets of miR-144-3p such as Smad4, FZD4 and TET2 [17-19]. The findings suggest that miR-144-3p plays a role in the pathogenesis of OA. However, the function of this miRNA molecule has not been completely revealed. To understand the function of miR-144-3p in OA, it is necessary to identify its target genes. Therefore, we used bioinformatics tools to predict the target genes of miR-144-3p. Next, we demonstrated that this gene was a direct target of miRNA-144-3p in vitro.
2. MATERIALS AND METHOD
miRmap (https://mirmap.ezlab.org/app/), MicroRNA Target Prediction Database - miRDB (http://mirdb.org/), and TargetScanHuman (http://www.targetscan.org/vert_72/) were utilized to predict miR-144-3p potential targets. The sequences of mature hsa-miR-144-3p (miR-144-3p) were obtained from the miRBase database. The sequence of miR-144-3p binding sites were identified as “TACTGT” (6 mer), “ATACTGT” (7 mer-m8), “TACTGTA” (7 mer-A1) and “ATACTGTA” (8 mer).
SW1353 chondrocytes were cultured in DMEM media (Thermo Fisher) supplemented with 100 μg/ml of penicillin and streptomycin (Thermo Fisher), and 10% (v/v) fetal bovine serum (Thermo Fisher), at 37°C, 5% (v/v) CO2 until the cell growth reached 80% confluency. The cells were then seeded at a density of 2x105 cells/mL in a 6-well plate and cultured to 80% confluency. Cells were then transfected with 50 nM miR-144-3p mimics, miR-144-3p inhibitor (Qiagen) or corresponding control (Qiagen) using Lipofectamine 2000 (Thermo Fisher) for approximately 24 hours according to the manufacturer’s protocol.
Total RNA from cells was extracted using TRIzol Reagent (Invitrogen) and converted to cDNA using the SuperScript II RT kit (Thermo Fisher). The concentration and purity of extracted total RNA were checked by Nanodrop using A260/280 and A260/230 ratios. Expression of ADAMTSL3 was determined using the SensiFAST SYBR No-ROX Kit (Bioline), forward primer 5’-TGGAAGCATGTGATGAGAGC-3’ and reverse primer 5’-TTGACTGTCTGCTGGGTCTG-3’ with an internal control, 18S rRNA. Expression of ADAMTSL3 was determined using the ΔCT formula. Realtime-PCR quantification was performed using a LightCycler® 96 System (Roche).
The 3’UTR fragment with the miR-144-3p-binding sites of ADAMTSL3 was cloned into pmiRGlo vector by the In-Fusion HD Cloning kit (Takara). The forward primer 5’-GCTCGCTAGCCTCGATTAGGTCAGGGTTGGATTGG-3 ’ and reverse primer 5’-CGACTCTAGACTCGAGGATCATGAAACAGAAACAT TAACTC-3’. The mutated pmiRGlo-ADAMTSL3 vector was kindly provided by an overseas research group. The cloning product was checked by colony PCR using forward primer 5’-TTCACTGACACTGGGTTGAGCACTACTG-3’ and reverse primer 5’-CGACTCTAGACTCGAGGATCATGAAACAGAAACATTAACTC-3’, and subsequently sequenced.
SW1353 cells were seeded in a 96-well plate at a density of 5x104 cells/mL in 100 μL of medium and cultured overnight. The plasmids including pmiRGlo-ADAMTSL3 (100 ng), mutant pmiRGlo-ADAMTSL3 (100 ng), miR-144-3p mimics (50 nM ) and a control (50 nM ) were then transfected seperately using Lipofectamine 2000 (Thermo Fisher). The level of luciferase activities was determined using the Dual-luciferase Reporter kit (Promega) and detected using an EnVision 2103 Multilable (Perkin Elmer). Normalized luciferase activity was reported as the Firefly luciferase/Renilla luciferse activity.
3. RESULTS
Among potential target genes of miR-144-3p predicted by bioinformatics algorithms , we selected candidates that have been reported to be associated with OA in previous studies. TargetScanHuman and miRDB predicted that miR-144-3p had round 1043 and 1254 target genes, respectively. The difference in the number of target genes proposed by two bioinformatics tools is probably due to the discrepancy in predictive analytics algorithms.
Of these predicted target genes, ADAMTSL3 was identified by both softwares with two seed sites of miR-144-3p on the 3’UTR (Figure 1). TargetScan results showed that ADAMTSL3 has two 7mer-A1 (TACTGTA) binding sites for miR-144-3p, of which position 1593-1599 is highly conserved (Figure 1A) and position 1926-1932 is poorly conserved (Figure 1B). Similarly, the miRDB tool also revealed two 7mer-A1 (TACTGTA) sites for miR-144-3p on the ADAMTSL3 3’UTR were located at position 1593 and 1926 (Figure 1C). These data proposed that miR-144-3p possibly binds to the 3’UTR of ADAMTSL3.

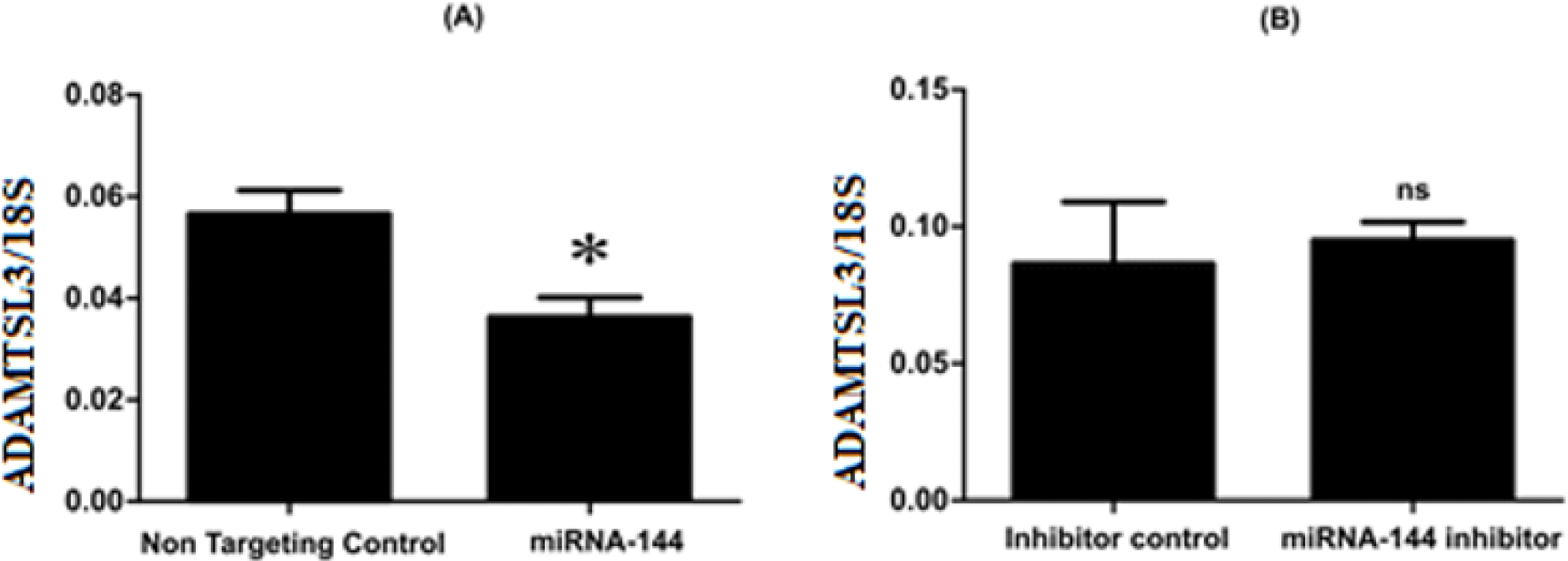
The expressions of ADAMTSL3 in SW1353 cells after being transfected with miR-144-3p mimics or miRNA-144-3p inhibitor and control were observed using Realtime-PCR. Upon miR-144-3p overexpression, there is a reduce in ADAMTSL3 expression compared to non-targeting control (Figure 2A). Meanwhile, the inhibition of miRNA-144-3p had insignificant effect on the expression level of ADAMTSL3 compared to control (Figure 2B).
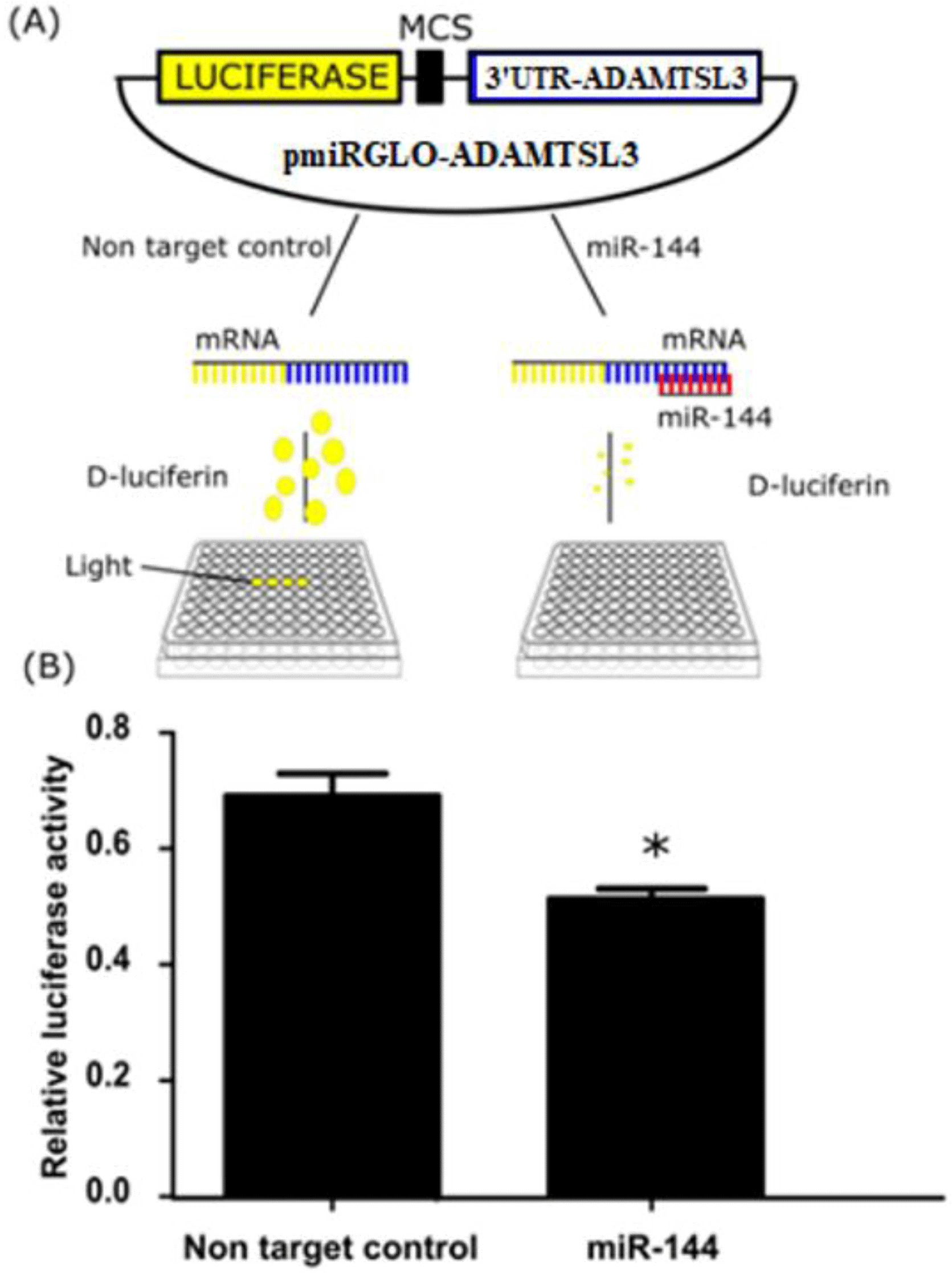
To investigate whether miR-144-3p directly interacts with ADAMTSL3 at the binding sites, the 3’UTR of ADAMTSL3 containing the miR-144-3p-binding was inserted downstream of the Firefly luciferase reporter gene on the pmiRGlo vector. The pmiRGlo-ADAMTSL3 was co-transfected with miR-144-3p mimics into SW1353 cells which were subsequently cultured for 24 hours before measuring luciferase luminescence expression. As expected, the luciferase activity reduced compared with non-target control (Figure 3).
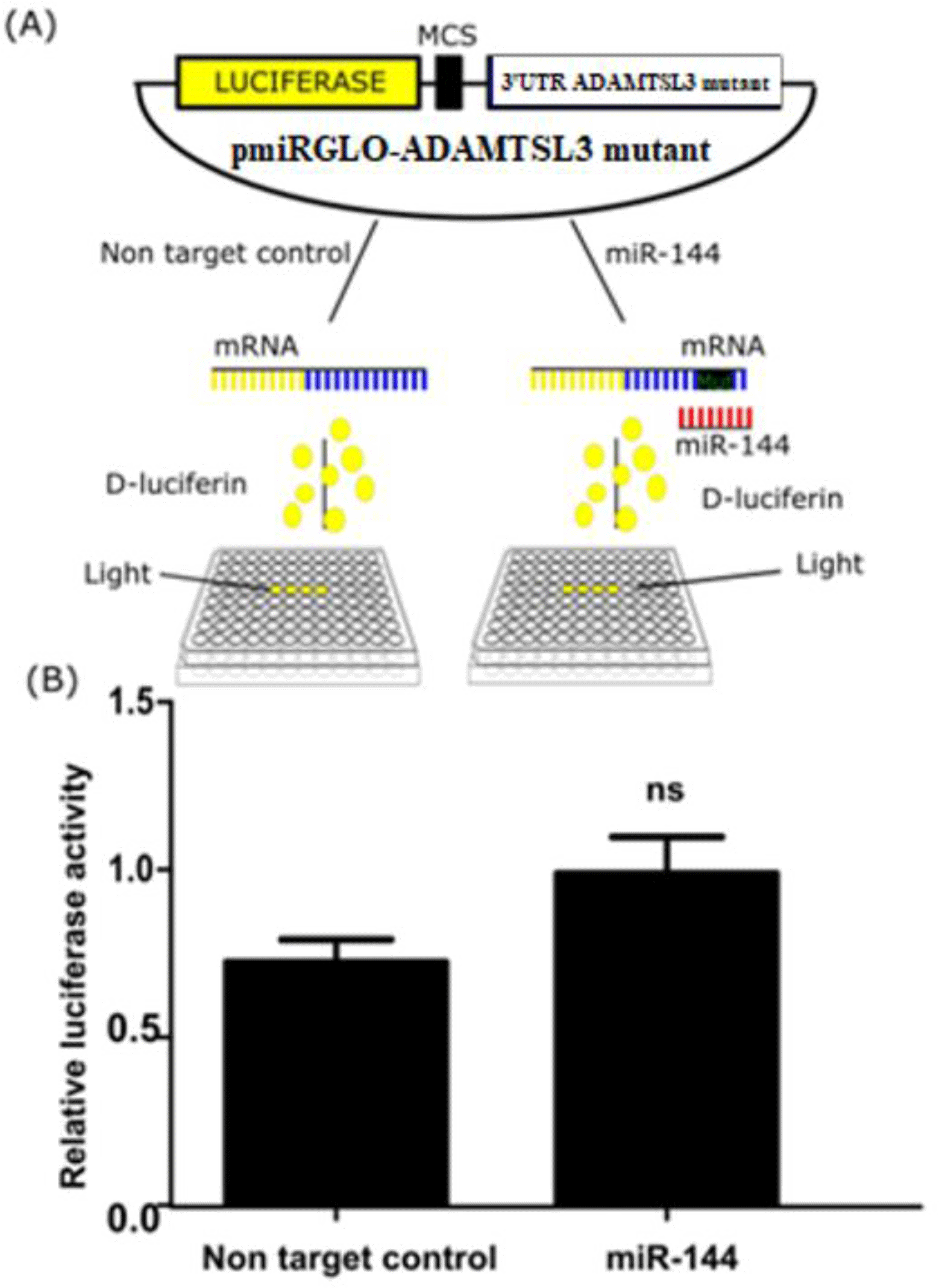
Next, a similar experiment was conducted using mutant pmiRGlo-ADAMTSL3 harboring the two mutants at seed binding sites of miR-144-3p. The mutant vector (100ng) was co-transfected with miR-144-3p mimics or non targeting control for 24 hours and again measured luciferase activity. The relative luciferase activity was recovered compared with controls (Figure 4).
4. DISCUSSION
We previously identified miR-144-3p overexpression in OA disease [15]. We next determined the function of this molecule by identifying its direct targets by employing various bioinformatics tools and then validating the prediction in vitro. The results indicated that ADAMTSL3 is the direct target of miR-144-3p.
The data of the in silico investigation have identified ADAMTSL3 as a potential target gene of miR-144-3p because on the 3’UTR of the mRNA, there are two miR-144-3p binding sites whose functions are related to OA disease [20]. The novel finding demonstrated that ADAMTSL3 is the target gene of miR-144-3p in OA. Moreover, we found that the expression of ADAMTSL3 was affected by the abundance of miR-144-3p in the cells. In particular, upon the accumulation of miR-144-3p, the expression of ADAMTSL3 decreased. In contrast, upon the depletion of miR-144-3p, ADAMTSL3 marginally increased (not statistically significant). Taken together, the data suggest that miR-144-3p inhibits the expression of ADAMTSL3. However, this result could not determine whether miR-144-3p directly or indirectly (via another molecule) regulates ADAMTSL3. It is necessary to conduct further experiments to answer this question. We also observed an increase of ADAMTSL3 when miR-144-3p in the cells was inhibited, but it was not statistically significant because the endogenous miR-144-3p at the time of the investigation was quite low, which may cause the upregulation of ADAMTSL3.
In addition, we found that when binding sites for miR-144-3p was cloned into the 3’UTR region of the luciferase gene, the activity of reporter gene decreased. The activity was, however, restored when we introduced mutation in the binding sites of miR-144-3p. These results indicated that miR-144-3p acts directly on the ADAMTSL3 mRNA through its binding site on the 3’UTR.
Conclusion
ADAMTSL-3 (also known as punctin-2) is a member of the ADAMTSL family together with ADAMTSL-1, -2, and -3. ADAMTSL-3 is a glycoprotein whose molecular weight is 210-kDa. It possesses 13 TS repeats and is present in the extracellular matrix of various tissues [21]. The function of ADAMTSL-3 has not yet been fully defined. It has been reported to involve both in cell-extracellular matrix interactions and cell-cell interactions as well as control the function of ADAMTS proteases [21].
The increased miR-144-3p in OA was observed previously [15], and the current finding suggests that miR-144-3p may have an adverse interaction with ADAMTSL3. The disregulation of miR-144-3p could lead to the abbrent in the interaction between chondrocyte cells and extracellular matrix. This may consequently lead to OA. However, further studies need to be done before clinical applications.









