1. INTRODUCTION
Colorectal cancer (CRC) is one of the most common can-cers worldwide, ranking third in incidence and second in cancer-related mortality [1]. Lynch syndrome (LS), also known as hereditary nonpolyposis colorectal cancer (HNPCC), is an autosomal dominant genetic disorder caused by pathogenic germline mutations in DNA mismatch repair (MMR) genes, primarily MLH1, MSH2, MSH6, PMS2, and EPCAM [2,3]. LS is the most common hereditary cause of CRC, accounting for approximately 2%–4% of all CRC cases [4–6]. LS is an autosomal dominant genetic disorder characterized by an increased risk of developing CRC as well as several other cancers, such as endometrial, gastric, ovarian, and small intestine cancers [2,7,8].
In the context of healthcare in Vietnam, the annual incidence of CRC has been significantly increasing [9,10]. However, the literature body on the prevalence and characteristics of CRC in patients with LS remains limited. This may be due to the complex and specialized genetic tests required to diagnose LS, which may not be available in all healthcare facilities. Therefore, specific research on LS in Vietnam is essential to better understand the prevalence and clinical features of affected patients, and to develop effective prevention, diagnostic, and treatment measures.
The objectives of this study were to estimate the prevalence and to describe the clinical characteristics of LS among CRC patients in a hospital-based population in Vietnam. By analysing data from patients diagnosed and treated at University Medical Center of Ho Chi Minh City from 2022 to 2024, this study aimed to provide an overview of the current situation, thereby contributing to improve diagnostic and therapeutic quality for CRC patients with LS in Vietnam.
2. MATERIALS AND METHODS
This is a cross-sectional study on prospective data at the University Medical Center-Ho Chi Minh City, from 2022 to 2024.
Inclusion criteria: Patients selected for the study must meet the following criteria: (1) diagnosed with CRC; (2) presented with complete medical records and necessary clinical information for the study; (3) agreed to undergo the Mencare genetic panel test (including genes related to LS: MLH1, MSH2, MSH6, PMS2, and EPCAM) to evaluate the possibility of having LS.
Exclusion criteria: (1) patients whose postoperative pathology did not confirm CRC; (2) patients with incomplete medical records or missing important information for the study; patients with Crohn’s disease, ulcerative colitis, or familial adenomatous polyposis.
A review of the literature indicated that the prevalence of LS in CRC patients ranged from 1% to 5% [4]. We hypothesized that the prevalence rate was 2%. We aimed to estimate the prevalence of LS such that the estimate could not deviate more than 2% from the true population prevalence. We used the standard formula for estimating a single population proportion:
-
n = required sample size
-
p = estimated population proportion (prevalence rate is 2%)
-
m = margin of error (0.02 for ±2%)
-
Z1−∝/2 = 1.96 (corresponding to a 95% confidence level)
Based on the calculation, the required sample size to estimate the prevalence of LS that does not deviate more than 2% from the true population prevalence was 189 patients.
DNA samples were extracted from peripheral blood from all participants. After collection at the hospital, blood samples were sent to GeneSolutions company for germline testing using the Mencare panel.
Genomic DNA was extracted from blood cells using the Illustra Blood GenomicPrep Mini Spin Kit (GE Healthcare®, Marlborough, MA, USA) according to the manufacturer’s protocol. The DNA concentration and purity were assessed using a QFX Fluorometer (DeNovix®, Wilmington, DE, USA). The minimum required DNA concentration was 2.5 ng/μL to ensure sufficient quality for downstream applications.
Primers were designed to amplify the coding regions of 10 target genes (MLH1, MSH2, MSH6, PMS2, EPCAM, APC, MUTYH, CDH1, BRCA1, and BRCA2). The primers were validated based on the following criteria: (1) maximum amplicon size: 140 bp; and (2) coverage: ≥95% of the targeted regions. Primer design followed AmpliSeq Gene principles and was performed using Design Studio software (https://www.illumina.com/products/by-type/informatics-products/designstudio.html). The primers were synthesized by Illumina® (San Diego, CA, USA) and organized into three master mixes (Pool 1, Pool 2, and Pool 3).
Genomic DNA extracted from blood samples was amplified using a multiplex-PCR reaction with three pools of primers. The PCR products (amplicons) were treated with FuPa reagent to remove excess primers, and adapters were added to uniquely index each sample. The DNA library was purified by AMPure XP beads prior to amplification. The final library concentration was quantified using a QFX Fluorometer, followed by dilution to a final concentration of 2 nM.
NGS was performed using the MiniSeq system (Illumina®) with a MiniSeq High Output Kit. The number of samples per run was calculated to ensure a sequencing depth of 600–1,000× coverage for each sample.
NGS data were analysed using BaseSpace Sequence Hub software (Illumina®), with human genome GRCh19 as the reference. Variants were classified using ClinVar database for germline mutations, and Catalogue of Somatic Mutations in Cancer (COSMIC) database for known cancer-associated variants.
All genetic variants identified by NGS were subsequently validated using direct Sanger sequencing with appropriate primers. The confirmation protocol followed previously established methods. Germline variants were reported following the Human Genome Variation Society (HGVS) guidelines. The functional impact of novel missense mutations was predicted using Sorting Intolerant From Tolerant (SIFT) and Polymorphism Phenotyping v2 (PolyPhen-2).
Data were collected from the medical records of patients at the University Medical Center-Ho Chi Minh City. The information collected included:
-
Personal Information: Age, gender, BMI, and family history of cancer. The population was divided into two groups, namely <50 years and ≥50 years, based on established screening criteria for LS and early-onset CRC [2,3].
-
Clinical Characteristics: Initial symptoms, tumour location, cancer stage (according to the TNM system), and relevant tests.
-
Genetic Testing: Results of the Mencare genetic panel test (including genes related to LS: MLH1, MSH2, MSH6, PMS2, and EPCAM) to evaluate the possibility of having Lynch syndrome.
-
Treatment and outcomes: Treatment methods (surgery, chemotherapy, radiotherapy) and treatment outcomes.
Data were analysed using SPSS software version 25.0. Before applying statistical tests, data distribution was assessed to ensure the appropriate choice of methods. Normality of continuous variables was assessed using the Shapiro-Wilk test. For variables that did not meet these assumptions, the non-parametric Mann-Whitney U test was used. Statistical analyses included:
-
Proportion Analysis: The proportion of patients with LS in the total number of CRC patients.
-
Descriptive Statistics: Used to describe the general characteristics of the study sample, including frequency, percentage, mean, and standard deviation.
-
The 95% confidence intervals (CIs) for prevalence estimates were calculated using the exact binomial (Clopper-Pearson) method.
-
Chi-square Test (χ² test): Used to compare categorical variables between patients with and without LS when expected frequencies were sufficient.
-
Fisher’s Exact Test: Applied to categorical variables when more than 20% of the expected cell counts were less than 5, to ensure statistical validity in analyses involving small sample sizes.
-
Independent t-test: Used for comparing means of normally distributed continuous variables between LS and non-LS groups.
-
Mann-Whitney U test: Non-normally distributed continuous variables were presented as median and interquartile range (IQR) and compared using the Mann-Whitney U test.
The protocol of the study was approved by the Institutional Review Board (IRB) of University of Medicine and Pharmacy at Ho Chi Minh City, Vietnam under Decision no. 618/HĐĐĐ-ĐHYD. All personal information of the patients was kept confidential and used only for research purposes. Participants signed an informed consent form after being clearly informed about the study’s objectives and content.
This study was conducted and reported in accordance with the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines to ensure comprehensive and transparent reporting of observational research [11].
3. RESULTS
From March 2022 to March 2024, a total of 204 patients provided consent to participate in the study and satisfied the inclusion criteria. Postoperative pathological analysis revealed that 14 patients were not diagnosed with carcinoma. As a result, a total of 190 patients were included in the final data analysis.
The demographic and clinical characteristics of the population are summarized in Table 1.
The overall prevalence of LS in this cohort was 12 out of 190 patients, corresponding to 6.3% (CI 95%: 2.85%–9.75%). The prevalence of common mutations (MLH1, MSH2, MSH6) was found in 7 out of 190 patients, corresponding to 3.7% (CI 95%: 1.02%–6.38%).
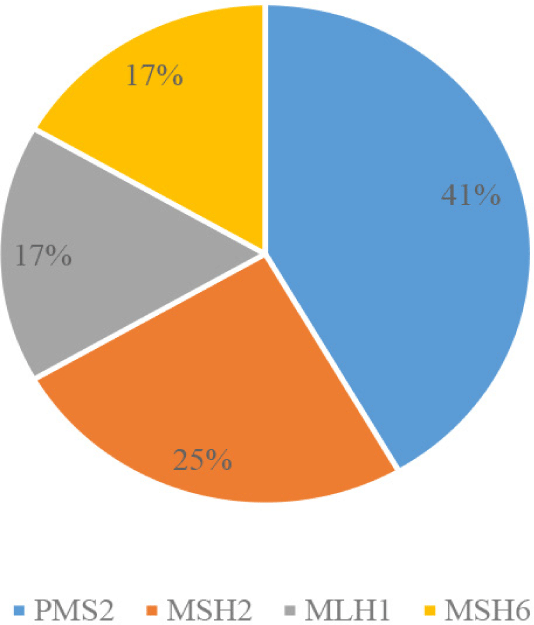
The analysis of the 12 patients diagnosed with LS revealed significant insights when examining the data by gene mutations, age groups, tumour location, cancer stage, and pathology. In terms of gene mutations (Fig. 1), PMS2 is the most frequently observed, presented in 5 out of 12 patients (41.7%), followed by MSH2 (33.3%), MLH1 (16.7%), and MSH6 (16.7%).
This suggested a higher prevalence of PMS2 mutations than typically reported, emphasizing the importance of including this gene in genetic screenings.
Regarding family history, 7 out of 12 LS patients (58.3%) reported no family history of CRC. By age groups, 6 patients (50%) were diagnosed at the age of 50 or younger, and 6 were diagnosed at 50-year-old or above. In the younger group (30 to 49 years), mutations in PMS2, MSH2, MLH1, and MSH6 were present, highlighting the early onset potential of LS. In the older group (53 to 73 years), PMS2 and MSH2 mutations continued to be prominent, with 3 out of 6 patients having PMS2 mutations, suggesting the significance of this mutation even in older age.
Gender distribution was balanced with 6 males and 6 females, indicating no significant gender predisposition. Tumour location was most common in the right colon (58.3%), followed by the left colon (25%) and rectum (16.7%), aligning with the typical pattern in LS. The cancer stage analysis showed that half of the patients were diagnosed at stage 3, indicating a tendency toward late-stage diagnosis. Pathologically, the majority of tumours were moderately differentiated (75%), with a few poorly differentiated or mucinous carcinomas, which are often associated with more aggressive disease.
The characteristics of the study population are summarized in Table 2. The study included 99 males (52.1%) and 91 females (47.9%). Among the LS positive group, there were 5 males (5.1%) and 7 females (7.7%). There was no statistically significant difference in gender distribution between LS positive and LS negative groups (p=0.455).
The mean age of the study population was 59.6±12.4 years. The mean age in the LS positive group was significantly lower at 49.7±14.5 years compared to 60.3±12.1 years in the LS negative group (p=0.004). Patients were categorized into two age groups: <50 years and ≥50 years. There were 41 patients (21.6%) under 50 years old and 149 patients (78.4%) aged 50 years or older. Among the LS positive patients, 6 were under 50 years old (14.6%) and 6 were 50 years or older (4.0%), showing a significant difference (p=0.024).
The mean body mass index (BMI) of the overall study population was 22.53±3.33 kg/m². In group comparison, the LS-positive patients had a lower BMI mean (21.47±1.13 kg/m²) compared to LS-negative patients (22.59±0.25 kg/m²). However, this difference was not statistically significant (p=0.255).
There were no significant differences in the use of neoadjuvant chemotherapy, the occurrence of intraoperative complications, or the methods and types of anastomoses between the LS positive and LS negative groups. The distribution of tumour location, staging, pathology types, tumour size, surgery duration, and blood loss did not show any statistically significant differences between groups.
There were no significant differences in the time to first bowel movement (2.78±1.09 days vs. 2.84±1.21 days, p=0.889) or length of hospital stay (6.78±0.83 days vs. 7.19±2.20 days, p=0.234) between the two groups.
4. Discussion
The prevalence of LS in our study cohort of CRC patients was 6.3%. When focusing on the key genes MLH1, MSH2, and MSH6, the prevalence was 3.7%, aligning closely with Nadine’s 2022 meta-analysis, which reported an average LS prevalence of 2.2% across 51 studies worldwide [4]. This meta-analysis found higher prevalence rates in studies employing germline testing (up to 5.1%) and lower rates in studies using initial MSI or IHC screening (around 1.1%) [4].
Previous studies in Northeast Asia reported lower LS prevalence. For instance, Jeong’s 2003 study in Korea found a prevalence of 0.4% among 230 CRC patients [12]. Chika’s 2017 study in Japan reported a 0.7% prevalence among 1,234 CRC patients [13]. Similarly, Yao’s 2021 study in Shandong, China, showed a prevalence of 0.6% among 1,294 patients [14]. These lower rates may reflect regional genetic differences and the methods used for screening.
However, more recent studies in other parts of China have reported higher prevalence rates. Dong’s 2020 study in Beijing found a 2.7% prevalence in a cohort of 4,195 CRC patients, and Jiang’s 2021 study in Guangzhou reported a prevalence of 2.9% among 3,330 patients [15,16]. These findings align more closely with our study and suggest regional variations within China.
Data on LS prevalence in CRC patients in Southeast Asia is limited. However, a study in Thailand reported a 3% prevalence of LS among endometrial cancer patients, suggesting potential similarities with the Vietnamese population [17]. In the Philippines, a study on young-onset CRC patients found that 21% had deficient mismatch repair (dMMR) status, with higher deficiency prevalence in MSH2 and MSH6 (9%) than MLH1 and PMS2 (5%) [18]. These findings indicate a significant presence of LS-related genetic mutations in the region.
The relatively high prevalence of LS observed in our study may be attributed to two key factors. First, we employed comprehensive germline genetic testing using a multigene panel that included MLH1, MSH2, MSH6, PMS2, and EPCAM. This approach enabled us to detect mutations in PMS2, a gene that is often underrepresented or even omitted in some screening protocols. Notably, PMS2 was the most frequently mutated gene in our LS-positive group (41.7%) and was known to be associated with lower penetrance and later onset, which might lead to underdiagnosis when using more selective testing strategies.
Second, our study included patients across a broad age spectrum, rather than restricting inclusion criteria to early-onset CRC cases or those met strict family history criteria. As a result, we identified 50% of LS cases diagnosed at the age of over 50-year-old and that 58.3% had no reported family history of CRC. These findings underscored the limitations of relying solely on age or family history as pre-screening criteria, which may have contributed to the lower prevalence estimation than those in previous studies with similar filters.
In combination, these methodological strengths—broad genetic panel testing and inclusive selection criteria—likely enhanced our ability to detect LS with better comprehensiveness, thereby contributing to the higher observed prevalence compared to studies using narrower testing scopes or selective enrolment strategies.
Our study identified a notable number of PMS2 mutations among CRC patients with LS. This finding contrasts with earlier studies that predominantly reported mutations in MLH1, MSH2, and MSH6. For example, Moreira’s extensive study in the USA identified 312 LS patients with mutation rates of 37% for MLH1, 41% for MSH2, 13% for MSH6, and only 9% for PMS2 [19]. Similarly, Dong’s study in China found 115 LS patients with 39% MLH1 mutations, 34% MSH2 mutations, 12% MSH6 mutations, 9% PMS2 mutations, and 5% EPCAM mutations [15].
The higher prevalence of PMS2 mutations in our study can be attributed to the comprehensive genetic screening that included PMS2, unlike many earlier studies that focused primarily on MLH1, MSH2, and MSH6 due to clinical criteria such as the Bethesda and Amsterdam guidelines. These guidelines tend to underrepresent PMS2 mutations because PMS2 is often associated with later-onset CRC, which may not be captured as effectively by criteria designed for early-onset cases [7].
Earlier studies that utilized these clinical criteria were likely to miss multiple PMS2 mutation carriers. For example, the PLSD study highlighted that PMS2 carriers did not show a significant increased risk of cancer before the age of 50-year-old, which indicated that they might be overlooked if the screening focuses on younger patients [7]. By including PMS2 in our genetic panel, we were able to detect a broader range of mutations, providing a more comprehensive picture of LS in our population.
The prevalence of PMS2 mutations in our study might also indicate genetic differences between the Vietnamese population and other populations studied. Genetic diversity among populations can lead to variations in mutation frequencies. Studies in different Asian populations have shown varying mutation distributions, suggesting that regional genetic factors may play a significant role [12,13,15,18].
Historically, screening strategies for LS have focused on younger patients, typically under 50-year-old, based on clinical criteria such as the Amsterdam and Bethesda guidelines. However, our findings highlighted the need to extend screening to older populations. Notably, in our study, four patients with PMS2 mutations were over the age of 50 (aged 57, 58, 66, and 73 years), and two patients with MSH2 mutations were also over 50-year-old (aged 65 and 73 years). This supported the argument for expanding LS screening to include older individuals.
Moreira’s study, which found that 55% of LS patients were over 50 years old, and Pearlman’s study, which reported a similar rate of 52%, both underscored the importance of including older patients in LS screening programs [19,20]. The study by Dong in Beijing and Jiang in Guangzhou also found significant proportions of over-50-year-old LS patients, at 49% and 42% respectively [15,16].
Extending the age limit for genetic testing to 70 years, as recommended by Sie’s study in the Netherlands, could significantly improve LS detection rates. Sie’s study demonstrated that this strategy could quadruple the detection rate of LS in CRC patients compared to screening only those under 50-year-old, making it both cost-effective and beneficial for the quality-of-life of mutation carriers [21].
Family history is a crucial component of LS screening. However, our study showed that a significant number of LS patients did not have a known family history of CRC. For example, seven out of 12 patients (58.3%) with LS in our study reported no family history of CRC. This highlighted the limitations of relying solely on family history for LS screening. Genetic mutations such as PMS2, which were prevalent in our study, might not be captured if family history is the primary screening criterion. Clinical guidelines like the Amsterdam and Bethesda criteria, which heavily weigh family history, may return false-negative cases in populations where family history is less reported or documented [3,7,22].
Our study features several strengths, notably the use of comprehensive genetic screening that included a broad panel of genes: MLH1, MSH2, MSH6, PMS2, and EPCAM. This approach allowed us to detect a wide range of mutations, including those in PMS2, which might have been missed by more traditional screening methods focusing only on MLH1 and MSH2. Another significant strength is the inclusion of patients across a wide age-range. This inclusion provided a more accurate picture of LS prevalence across different age groups, highlighting a significant number of LS cases in individuals over 50 years old. Furthermore, the detailed collection of clinical and genetic information for each patient, such as family history, tumour location, stage, pathology, and mutation type, offered a nuanced understanding of LS in the Vietnamese population.
However, our study also has notable limitations. The small sample size limited the generalizability of the findings, necessitating larger studies to confirm the prevalence and distribution of LS-related mutations in the Vietnamese population. Conducted at a single hospital, our findings may not be representative of the entire country, indicating a need for multi-centre studies to provide a comprehensive understanding of LS in various regions and healthcare settings. Additionally, the potential selection bias, where patients who agreed to genetic testing might differ from those who did not, could have influenced the observed prevalence rates. The study also faced challenges with incomplete family history data, which limited our ability to fully explore the impact of family history on LS detection. Finally, the lack of long-term follow-up data prevented us from understanding the prognosis and outcomes for patients with different LS-related mutations.
5. CONCLUSION
In conclusion, our study provided valuable insights into the prevalence and genetic mutation distribution of LS in the Vietnamese CRC patients. The comprehensive genetic screening and inclusion of a broad age range offered a significant strength, revealing a higher-than-expected prevalence of PMS2 mutations and underscoring the importance of inclusive screening practices. However, the study’s limitations, such as the small sample size, single-centre design, potential selection bias, and incomplete family history data, highlight the need for larger, multi-centre studies with long-term follow-up to enhance our understanding of LS. Addressing these limitations in future research will improve screening and management strategies for diverse populations, ultimately leading to better patient outcomes and more effective use of healthcare resources.