1. INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) including oral squamous cell carcinoma (OSCC) is a kind of cancer that affects the thyroid, salivary glands, oral cavity, and upper aerodigestive tract. [1]. The most prominent components of the tumor stroma are cancer-associated fibroblasts (CAFs). CAFs contribute to the formation and remodeling of the extracellular matrix (ECM) structure within the tumor microenvironment (TME) [2]. Partial epithelial-mesenchymal transition (p-EMT) playing a crucial role in invasion and metastasis is the intermediate state of epithelial-mesenchymal transition (EMT) during the evolution of epithelial malignancies when the cancer cells display both mesenchymal and epithelial characteristics [9]. CAFs and p-EMT tumor cells coexist and contribute to the growth of epithelial tumors such as HNSCC and OSCC.
We previously integrated 60,662 OSCC cells from GSE103322, GSE164690 and our own published datasets of single-cell RNA sequencing (scRNA-seq) [3-6]. This technology allows us to identify the tissue components, developmental hierarchies and immune patterns applicable to tumor biology and treatment [7]. From this integrated dataset, we sub-clustered 5,509 CAFs and p-EMT tumor cells including 2,270 non-(N0) and 3,239 lymph node metastasis cells (N1-Nx) [3]. We have identified the cell-cell communication of CAFs and p-EMT subcell types. Interestingly, we found the significantly increased Wnt (Wingless and Int-1) signaling pathway in metastasis condition. Wnt signaling pathways are a group of signal transduction pathways that influence developmental processes, cell proliferation, and tissue homeostasis. The canonical Wnt/β-catenin signaling pathway (β-catenin dependent), non-canonical Wnt/planar cell polarity (PCP) pathway, and non-canonical Wnt/calcium pathway have all been identified. All three pathways are activated by the binding of a Wnt-protein ligand to a Frizzled family receptor (FZDs), which delivers the biological signal into the cell. The non-canonical Wnt/PCP pathway controls the cytoskeleton and cell motility. The non-canonical Wnt/calcium pathway controls cellular calcium levels [8]. Wnt signaling pathway has previously been implicated in modifying cell behaviors such as proliferation, EMT induction, tumor growth, and as a rising targetable therapy for tumor therapy [9, 10]. However, most of the studies focused on canonical and less focused on non-canonical Wnt signaling pathways in OSCC CAFs [9, 11]. In the present study, we deeply analyzed Wnt signaling pathway (both canonical and non-canonical) using our previous single-cell RNA-seq data of OSCC CAFs and p-EMT tumor cells by computational methods. The study discovered the significance of the Wnt signaling pathway in OSCC CAFs and p-EMT tumor cells and proposed Wnt signaling indicators for prognosis.
2. MATERIALS AND METHOD
Integrated single-cell RNA-seq data of OSCC CAF and p-EMT clusters were obtained from our previous study [3]. These cells were from samples being primary tumor, oral cavity location, metastasis information and then analyzed by R Seurat v.4 package [12]. The Dotplot tool was used to show how feature expression varies between clusters. The size of a dot represents the proportion of cells in a cluster, and the color represents the scaled average gene expression. Using FeaturePlot, we plotted cells on a UMAP plot depending on gene expression. A similar approach was applied using Violin plot [13, 14].
The Python tool CellPhoneDB v3 was used to analyze cell-cell communication utilizing accessible receptors, ligands, and receptor/ligand interactions from scRNA-seq data (https://www.cellphonedb.org) [15]. Databases of CellPhoneDB include UniProt, Ensembl, PDB, IMEx consortium, and IUPHAR. From there, we filtered 43 available Wnt signaling interactions (WNT ligands and their receptors of both canonical and non-canonical Wnt signaling pathways) for our analysis.
Under non- and metastasis conditions, gene lists of canonical (REACTOME BETA CATENIN INDEPENDENT WNT SIGNALING-M27288) and non-canonical (GOBP NON-CANONICAL WNT SIGNALING PATHWAY-M10871) Wnt signaling pathways were processed for GSEA v.4.2.3 (https://www.gsea-msigdb.org) in CAF and p-EMT clusters of sing-cell RNA-seq data. GSEA was run with 1000 permutations for each of the gene sets and a nominal p-value of 0.05 for normalized enrichment score (NES) using Diff_of_Classes mode. This mode used the difference of class means to determine the fold change of log scale data) [16].
We applied HSCC data from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov) to perform survival studies based on gene expression levels (WNT5A, CTHRC1 and SFRP2) by GEPIA v2 method. For hypothesis assessment, the approach used the log-rank test. Cox proportional hazard ratio was applied in survival figures with 95% CI [17, 18].
3. RESULTS
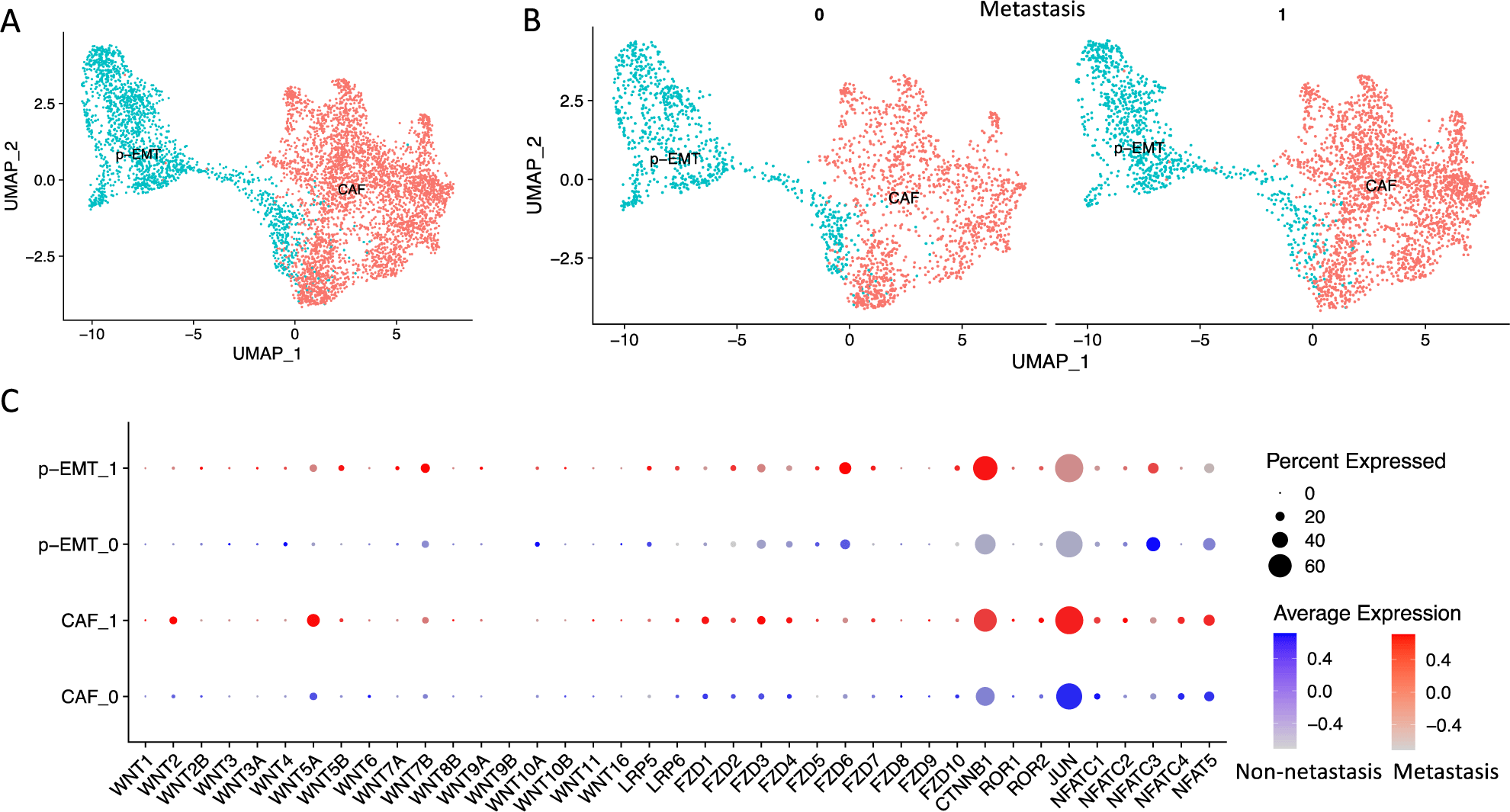
In the present study, we obtain the integrated scRNA-seq data of OSCC CAF and p-EMT clusters from our previous study [3]. These data contented 2,270 non-lymph node metastasis (N0) and 3,239 lymph node metastasis (N1-Nx) CAFs and p-EMT tumor cells (Figure 1A, B). We verified the expression of genes related to human Wnt signaling pathways including WNT ligands (WNT1, WNT2, WNT2B, WNT3, WNT3A, WNT4, WNT5A, WNT5B, WNT6, WNT7A, WNT7B, WNT8B, WNT9A, WNT9B, WNT10A, WNT10B, WNT11, WNT16), canonical co-receptors (LRP5, LRP6), receptors (FZD1, FZD2, FZD3, FZD4, FZD5, FZD6, FZD7, FZD8, FZD9, FZD10), canonical transcription factor (CTNNB1, β-catenin), non-canonical co-receptors (ROR1, ROR2), non-canonical transcription factors (JUN, NFATC1, NFATC2, NFATC3, NFATC4, NFAT5) (genes with no expression were not plotted) (Figure 1C). The results showed that WNT2, WNT5A, FZD1-4, LRP6, ROR2, NFATC1/2/4, NFAT5 increased in CAFs, while WNT7B, FZD2/5/6, LRP5/6, NFATC3 increased in p-EMT cells. These genes were also upregulated in metastasis vs. non-metastasis conditions. Significantly, both metastasis p-EMT and then CAF clusters strongly increased canonical transcription factor CTNNB1 (β-catenin) while both non- and metastasis CAF clusters increased non-canonical transcription factor JUN. These findings indicated that both CAF and p-EMT clusters increased genes related to Wnt signaling pathways. However, p-EMT tumor cells favored canonical Wnt signaling pathway (β-catenin dependent) and CAFs cells favored non-canonical Wnt signaling pathway (β-catenin independent). Metastasis condition increased these gene expressions.
Next, we investigated the Wnt signaling interactions (WNT ligands and their receptors/ co-receptors) of CAF and p-EMT clusters at the single-cell level using 43 Wnt signaling interactions (selected in CellPhoneDB database) (Figure 2A). In the non-metastasis condition, CAF/p-EMT expressed 5 different WNT interactions; CAF/CAF expressed 4 interactions; there was no interaction in p-EMT/p-EMT. In metastasis condition, CAF/p-EMT and CAF/CAF expressed 9 different WNT interactions; there was still no interaction in p-EMT/p-EMT. Figure 2B presented all 43 Wnt signaling interactions in CAF/CAF, CAF/p-EMT, p-EMT/CAF and p-EMT/p-EMT pairs. The results showed that most of the interactions were in CAF/CAF and CAF/p-EMT, especially in metastasis condition. In detail, there were 7 interactions not available in the non-metastasis condition (no gene expression). WNT5A ligand and its receptors co-receptors (FZD1/2/3/6, ROR1/2) interactions increased in CAF/CAF, CAF/p-EMT and p-EMT/CAF. These findings indicated that Wnt signaling interactions upregulated in CAFs and p-EMT cells, especially in metastasis condition.
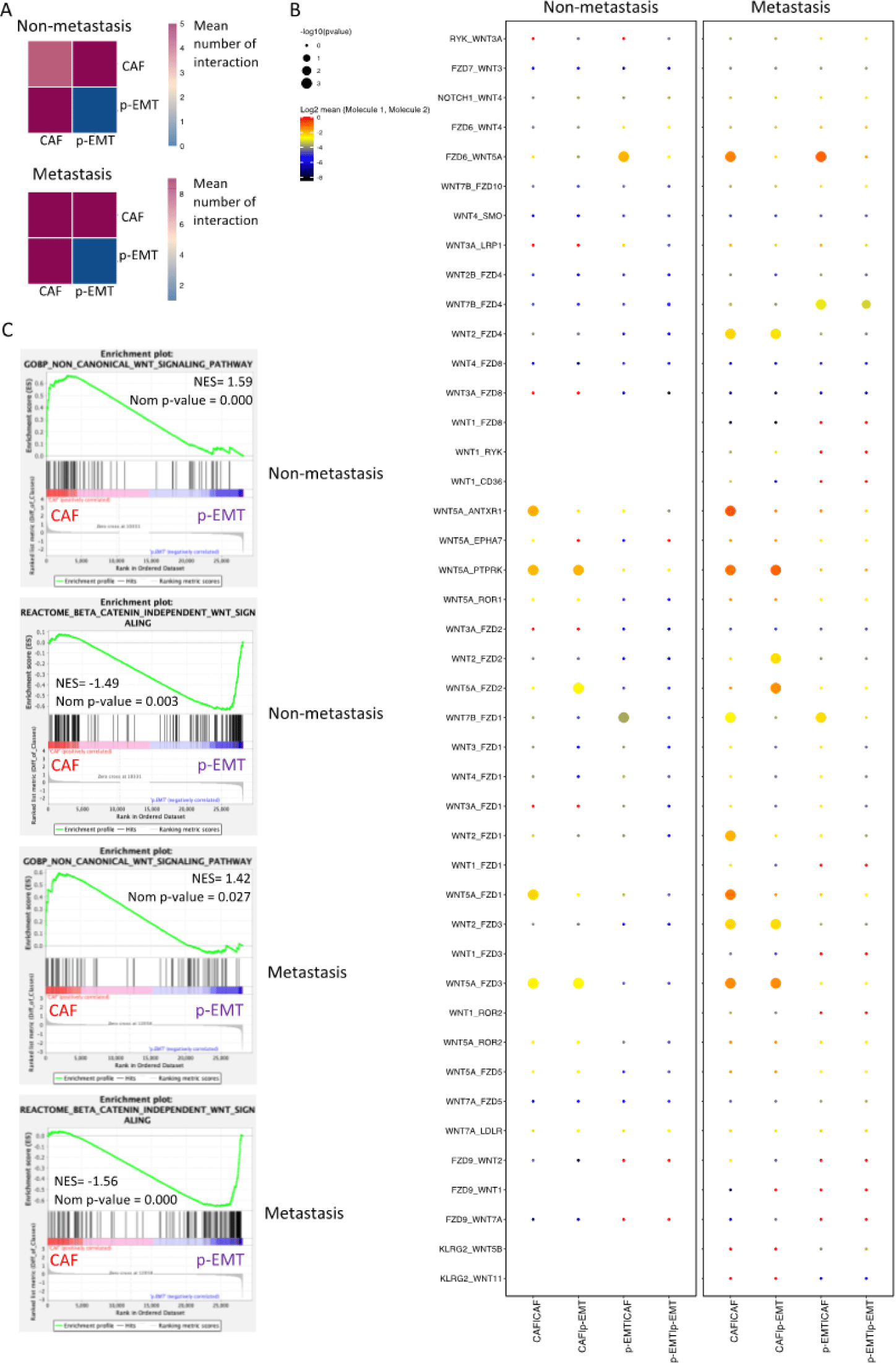
To determine the weights of the canonical and non-canonical Wnt signaling pathways in CAF or p-EMT clusters, we performed Gene Set Enrichment Analysis (GSEA) under non- and metastasis conditions. GSEA revealed the significant upregulation of the non-canonical Wnt signaling pathway in CAFs and canonical Wnt signaling pathway in p-EMT tumor cells in both non-vs. metastasis conditions (Figure 2C).
Considering the enhancement of Wnt signaling pathway in CAFs, especially the non-canonical one, we next checked the gene list’s expression of non-canonical Wnt signaling pathway in our cells in non- and metastasis conditions (Figure 3A). Genes were ranked by GSEA metric score from high expression to low expression with the top genes being WNT5A, CTHRC1 (Collagen triple helix repeat containing 1, an extracellular matrix Wnt binding protein of Wnt signaling pathway), SFRP2 (Secreted frizzled-related protein 2, a soluble modulator of Wnt signaling), SFRP4 (Secreted frizzled-related protein 4, a soluble modulator of Wnt signaling) in CAFs. These genes significantly contributed the highest parts in the enrichments of non-canonical Wnt signaling pathway in CAFs in both non-vs. metastasis conditions. Single-cell expressions of WNT5A, CTHRC1, SFRP2, SFRP4 in CAFs and p-EMT tumor cells in non- and metastasis conditions were illustrated by violin plots in Figure 3B. We found that WNT5A, CTHRC1, SFRP2 were highly upregulated in CAFs in metastasis condition. Co-expression plots of WNT5A/ CTHRC1 (Figure 3C), WNT5A/ SFRP2 (Figure 3D) at single-cell levels confirmed the upregulation of these genes in metastasis CAFs.
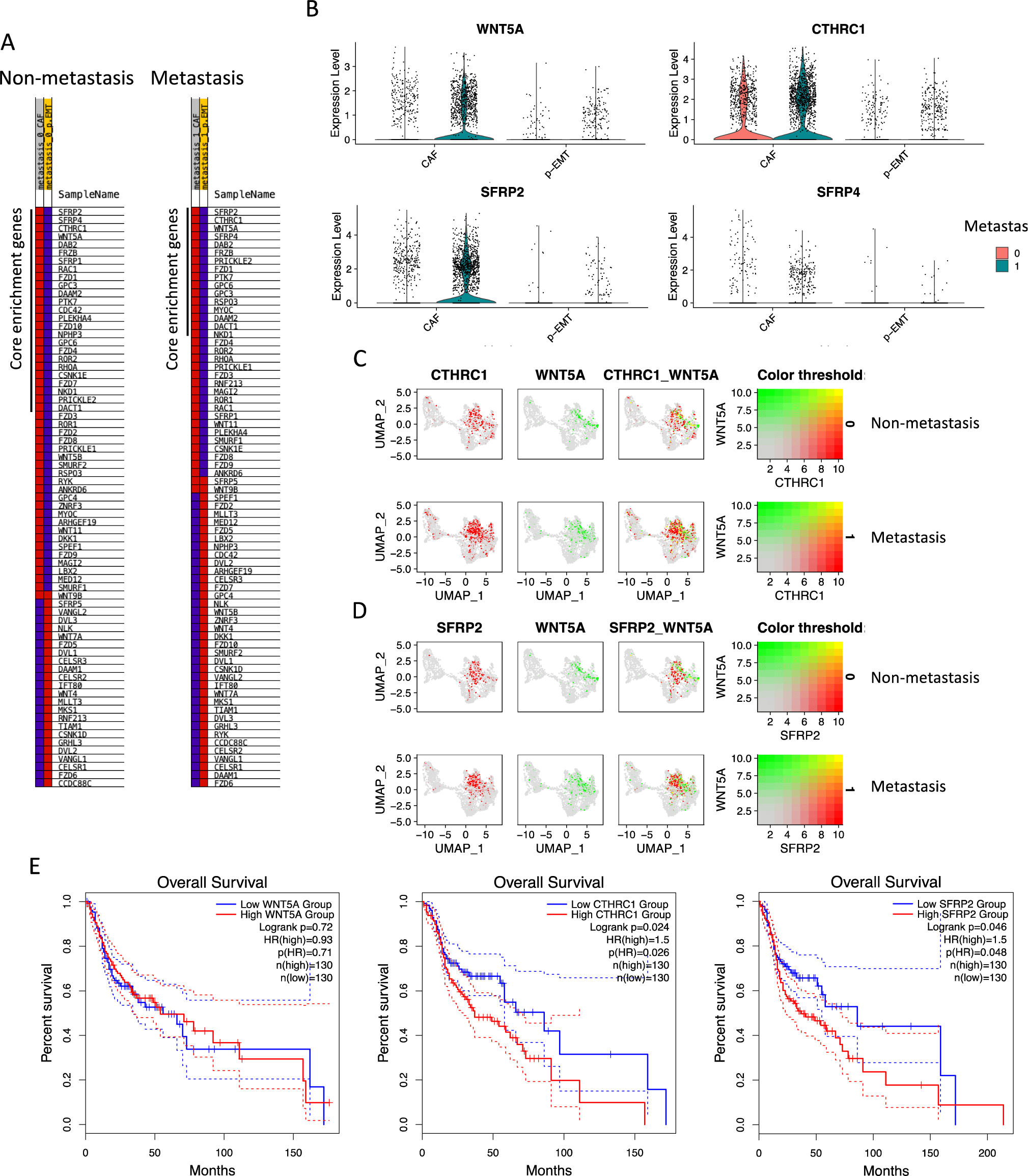
Finally, Overall Survival was analyzed using data of 260 HNSCC patients with high or low expression of WNT5A, CTHRC1, SFRP2 (n=130/group) using TCGA-HNSCC database (Figure 3E). Significantly, the outcomes of CTHRC1, SFRP2 groups were visualized by Kaplan-Meier plots. Patients with low expression of CTHRC1 were observed with 70% cumulative survival probability from 25 months up to 50 months, compared to 60% to 50% of the high expressing group. After 50 months, CTHRC1 low expressing groups maintained 70% cumulative survival probability up to 200 months. Remarkably, patients with low expression of CTHRC1 were observed with 30% cumulative survival probability in comparison to 10% of the low expressing group at 150 months. Similar results were found in SFRP2. After 150 months, these probabilities were 40% and 20% in low and high SFRP2 expression groups respectively. All the above findings indicated that CTHRC1 and SFRP2 are involved in Wnt signaling pathway and survival in HSCC and OSCC CAFs.
DISCUSSION
Current methods of scRNA-seq analysis allow understanding of cell-cell interaction through cell receptor– ligand networks [19, 20]. Our study highlighted the complex cellular communication via Wnt signaling pathway in OSCC CAFs and p-EMT tumor cells across metastasis conditions. We found that both CAFs and p-EMT tumor cells expressed strongly genes related to canonical Wnt signaling pathway, especially in metastasis condition. Cell communication also emphasized the WNTs and receptors/co-receptors in metastasis CAF/CAF and CAF/p-EMT. This finding indicated the upregulation of Wnt signaling in the ECM of TME.
In oral carcinogenesis, abnormal activation of the Wnt/β-catenin signaling pathway and progressive accumulation of nuclear β-catenin were prominent. It enhanced cell proliferation, migration and invasion in OSCC cells. High amounts of cyclin D1, surviving, and c-myc, which are typical Wnt/β-catenin targets, were found in OSCC showing that Wnt/β-catenin target genes may be elevated in oral cancer, leading to cell migration and invasion [9].
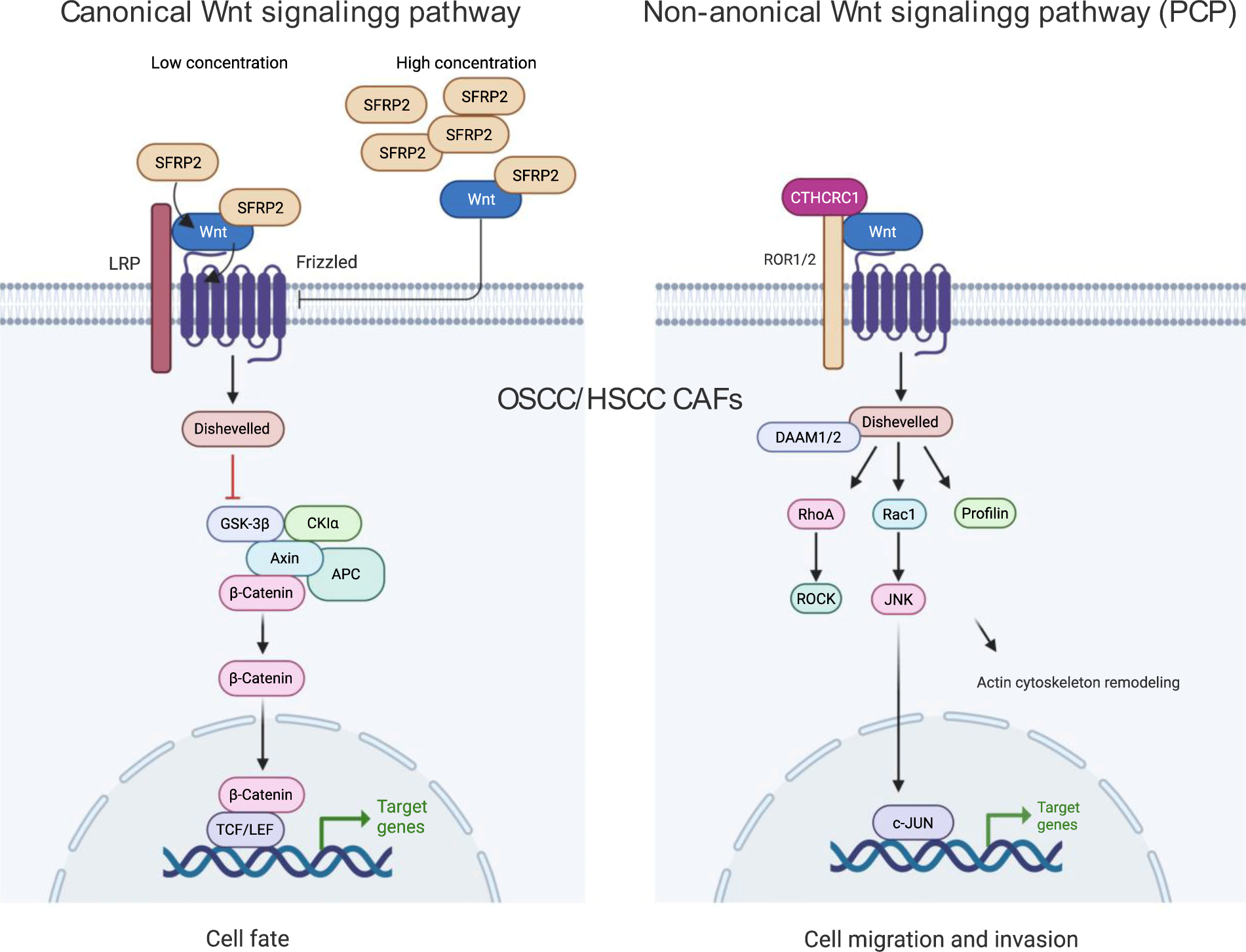
On the other hand, non-canonical Wnt signaling pathway was not investigated comprehensively in HSCC and OSCC. These β-catenin independent signaling pathways were divided into two pathways: Wnt/PCP signaling and Wnt/Ca2+ signaling, which is involved in cell motility, polarity, and migration [21, 22]. Studies in the lung, pancreatic and ovarian squamous cell carcinoma revealed the EMT promotion of Wnt/PCP signaling via WNT5A- and its non-canonical coreceptors ROR1/2 signal leading to tumor invasion and metastasis [23-25]. Significantly, in non-small cell lung cancer, WNT5A and ROR2 levels are related to a poor prognosis [26]. Additionally, Wnt/Ca2+ signaling was found in colon tumors and may promote epithelial morphological transformation via WNT11 signal. In the present study, we found a strong increase of Wnt/PCP signaling (c-Jun dependent) in CAFs, especially in metastasis condition.
A previous study showed that WNT5A has the potential to be a biomarker for the malignant transition of dysplasia to OSCC [27]. In our investigation, SFRP2 was highly expressed in OSCC CAFs and overall survival of HSCC suggesting a potential prognostic biomarker. This molecule belongs to the secreted frizzled-related protein (SFRP) family and acts as Wnt antagonist at high concentration and agonist at low concentration [28]. It was associated with metastasis of breast and melanoma cancer cells [28]. SFRP2 was found to be at a higher level in OSCC tumor tissue than in the margin clinically and could be a good prognostic factor of oral and gastric cancers [29-31]. Pointedly, we found the high expression of CTHRC1, an extracellular matrix Wnt binding protein of non-canonical Wnt signaling pathway in metastasis CAFs. CTHRC1 has been identified as a cancer-associated molecule where it promoted tumorigenesis, proliferation and invasion in many types of cancer [32]. In Wnt/β-catenin signaling pathway, CTHRC1 indirectly promotes the nuclear translocation and transcriptional activity of β-catenin to drive OSCC cell migration and invasion [33]. In Wnt/PCP non-canonical signaling pathway, CTHRC1 forms a Wnt receptor complex with FZD and ROR1/2 proteins to activate and transmit Wnt signals via Dvl-RhoA/Rac1-JNK-ATF2/c-Jun axis. This cascade then promotes tumor cell protrusion, proliferation, migration, and invasion [32]. CTHRC1 can be a prognostic biomarker of kidney carcinoma, breast cancer, colon adenocarcinoma and targeting this molecule may be beneficial for metastasis inhibition of non-small cell lung cancer [34-37]. Here we found that CTHRC1 was significantly associated with Wnt signaling pathway in OSCC CAFs and value biomarker for prognosis of disease survival (Figure 4). Last but not least, regarding the study’s limitations, the results of our targeted analysis should be validated by additional functional investigations on tumor cell lines and tissues as well as animal models of HSCC and OSCC in further studies.
Conclusion
In conclusion, our analysis highlighted the upregulation of Wnt/ β-catenin canonical and Wnt/PCP non-canonical signaling in CAF/CAF and CAF/p-EMT interactions at single-cell level, especially under metastasis condition. Our study suggests CTHRC1 and SFRP2 as biomarkers for HSCC and OSCC prognosis belong to CAFs of TME. It may lead to targetable therapy relying on WNT ligand-receptor interaction and Wnt signaling moderation in OSCC CAFs regarding the metastasis status.








