1. INTRODUCTION
According to WHO (2018), tooth decay is the most common non-communicable disease in the world, costs 5% of health care budgets and approx. 20% of out-of-pocket payments in developed countries, and is among the main causes for hospitalization of children in some high-income countries [1]. Dental plaque is considered a major cause that leads to tooth decay and is highly associated with mutans streptococci group. A systematic review conducted by N.L. Thenisch et al., (2006) summarized that a considerable increase in caries risk in young caries-free children was highly associated with the presence of mutans streptococci either in plaque or saliva [2]. T.M. Parisotto et al., (2010) also concluded in another systematic review that mutans streptococci levels are a strong risk indicator for early childhood caries [3].
When certain kind of carbohydrates, especially sucrose, are presenting in oral cavity, acidogenic mutans streptococci group (e.g. S. mutans, S. sobrinus…) could convert them into different types of insoluble exopolysaccharides (EPS) such as fructans and glucans. These EPS act like layers of biofilm that facilitate the adherence, colonization and accumulation of various anaerobic microorganisms such as Actinomyces spp., Fussobacterium spp., Spirochetes…[4, 5]. Acidogenic activities of these bacteria would lower pH of microenvironment inside plaque and that, in turn, corrodes and damages dental hard tissues and finally leads to dental caries. Streptococcus mutans is considered as a dominant producer of EPS in early stage of plaque development. Its strong aciduricity also helps S. mutans to survive in the acidic environment as well as outcompete against other oral bacteria [6, 7]. S. mutans produces three types of glucosyltransferase (gtfB, gtfC and gtfD) which are responsible for the production of α-1,3-glucan or α-1,6-glucan. Whereas gtfB and gtfC produce insoluble EPS, gtfD forms a soluble, readily metabolized polysaccharide and also acts as a primer for gtfB enhancing the overall synthesis of EPS [7, 8]. Aoki et al., (1986) reported that gtfB and gtfC were crucial for sucrose-dependent adhesion of S. mutans into substratum surface, while gtfD was not involved [9]. Moreover, gtfB was upregulated in biofilm cells relative to planktonics, and the transcription of gtfC could be also initiated from gtfB promoter [10]. In other words, gtfB is considered as the most important gene in EPS synthesis of S. mutans. In addition, cell to cell interactions in plaque multimicrobial communities, either inter- or intraspecies, also play a rule in late stage of dental plaque development. S. mutans expresses luxS which catalyses the reaction for the formation of autoinducer 2 (AI-2), a universally recognized signal by many bacteria species, which, in turn, maybe used to mediate interspecies interactions in multimicrobial communities [11]. LuxS also regulates different signaling pathways that affect the biofilm maturation, bacteriocin production and acid stress adaption [12, 13].
It is undeniable that S. mutans plays as a central role in dental plaque as well as dental caries development. Therefore, any therapeutic procedure that targets to suppress S. mutans growth and its biofilm formation would help to maintain good oral health condition. Nowadays, besides fluoride therapies, the use of probiotic for replacement or displacement of cariogenic bacteria in the oral cavity is a promising approach in the prevention of caries. Probiotics are defined by FAO/WHO (2001) as living microorganisms that, when administered in adequate amounts, confer health benefits to the host. Most of the probiotic bacteria used today are lactic acid bacteria (LAB) and are belong to the genera Lactobacillus or Bifidobacterium. Recent studies found that Lactobacillus spp. exhibit potential properties that could make them become a promising tool to fight against S. mutans and its relating diseases. Many of Lactobacillus spp. such as L. paracasei, L. rhamnosus, L. lactis, L. fermentum, L. reuteri, L. plantarum… were found to show an antagonistic activity against S. mutans by competing for adherence sites and nutrients; producing sort of antimicrobial secondary metabolites to inhibit S. mutans growth; and down regulating responsible genes in biofilm formation [14-23]. Some Lactobacilli could form specific co-aggregation with S. mutans thus confer the ease to remove these bacteria from oral cavity by swallowing [16].
Thus, in order to apply the benefits of these bacteria in daily dental caring, the aim of this current study was to isolate LAB exhibiting the ability to inhibit the growth and biofilm formation of S. mutans. Based on obtained results, oral functional foods such as lozenges or chewing gum containing these strains would be developed to aid in preventing dental plaque and tooth decay.
2. MATERIALS AND METHOD
Streptococcus mutans type strain ATCC 35668 was used as a pathogenic strain in in vitro antibacterial activity assay and biofilm formation assays. Bacterial colonies were obtained from -80°C glycerol stock by streaking onto Brain Heart Infusion (BHI) agar plate (Merck Millipore, USA) and incubating anaerobically at 37°C overnight. Inoculate one colony from this plate into Brain Heart Infusion Broth (BHB) media (Merck Millipore, USA) and incubate 16–18 h at 37°C. Dilute the culture in BHB until its optical density at 600 nm (OD600) reaches 0.08 – 0.1 (approx. 108 CFU/ml). The bacterial suspension was used within 15 minutes after being prepared.
Lactobacillus acidophilus type strain ATCC 4356 was used as a reference strain for biochemical tests, antibiotic susceptibility tests and 16S rRNA analyses of Lactobacillus spp. strain isolated in this study. All LAB (reference strain and isolated strains) were streaked onto De Man, Rogosa and Sharpe (MRS) agar (Merck Millipore, USA) and incubated anaerobically 24 – 48 h at 37°C. Inoculate one colony from these plates into MRS broth (Merck Millipore, USA) and incubate 16 –18 h at 37°C. Dilute the culture in sterilised phosphate-buffered saline (PBS) until its OD600 reaches 0.08 – 0.1 (approx. 108 CFU/ml). In antibiotic susceptibility test, transfer some colonies of LAB from MRS agar to Cation-adjusted Muller Hinton Broth (CAMB) until its OD600 is between 0.08 – 0.1. The bacterial suspension was continued diluting in CAMB supplemented with 5% lysed horse blood to get cell density of approx. 106 CFU/ml and was used within 15 minutes after being prepared.
Lactic acid bacteria were isolated from fermented vegetables and healthy human saliva samples. Eight types of vegetables (as shown in Table 1.) were collected from local markets and washed with distilled water then let dry in cool places. After being cut into small pieces, 0.5 kg of each type of vegetable was submerged in a clean glass jar containing approx. 2 liters of ferment solution (consisting of 20 g/l of glucose and 60 g/l of sodium chloride). After incubating these jars at room temperature in 5 – 7 days (as pH reaches 3.2 to 4.5), the fermented solution from these jars were harvested and used for isolating of lactic acid bacteria.
For human samples, saliva was obtained from students from our department. After being informed about the study, volunteers would be checked for not having tooth decays, tooth plaque and relating oral cavity diseases. Volunteers were then asked for not using foods, and alcoholic drinks as well as washing mouth with detergent agent 2 hours before sampling. Sterile cotton swabs were used to absorb saliva fluid from volunteers. Bacteria from these swabs were dispersed into 5 ml of sterile PBS tubes and were kept at 4°C no more than 2 hours till the bacteria isolation.
In short, samples (fermented vegetables and human saliva) were serial diluted and spread onto MRS agar plate supplemented with 5 g/L of calcium carbonate. After incubating anaerobically 24 – 48h at 37°C, colonies of acid producing bacteria (identified by a clear zone around its colony) were selected and restreaked onto MRS agar plate. LAB were identified on the basis of colony morphology (2-5 micrometers, convex, entire, opaque, and without pigment), gram staining (non-spore forming, rod-shaped gram positive bacteria) and basic biochemical tests (catalase-positive and oxidase-negative). Colonies defined as LAB were preserved in 20% glycerol stock at -80°C.
The growth inhibitory effect of LAB on S. mutans was The growth inhibitory effect of LAB on S. mutans was evaluated using the agar well diffusion method [24]. Preparation of culture supernatant from LAB: Inoculate a colony of LAB into 10 ml of MRS broth and incubate in 48 h at 37°C. Bacterial culture was centrifuged at 12,000 g for 10 mins at 4°C. The culture supernatant was passed through a 0.45 μm filter and adjusted to pH 7.0 using 1 N NaOH solution, subsequently.
S. mutans suspension prepared as described in section 2.1 was spread onto BHI agar plate using an aseptic cotton swab. Subsequently, wells of 6 mm diameter were punched into the agar and filled with 100 μl of LAB supernatant. Ampicillin (50 μg/ml) and sterilised MRS broth were used as positive control and negative control, respectively. After incubation, the diameter of inhibition zones were measured in mm. The assay was replicated three times with each Lactobacilli strain and data were expressed as mean (of inhibition zones) ± standard deviation.
Isolated LAB that showed the inhibitory effect on S. mutans growth were examined for the ability to reduce biofilm formation by colorimetric assay, described by O’Toole et al., (2011) with some modifications [25]. Briefly, a colony of either tested LAB or S. mutans was inoculated into BHB media at 37°C overnight. The bacterial cultures were diluted by BHB media until their OD600 nm was between 0.08 – 0.1. The 24-well plate was used to test the effect of LAB on S. mutans biofilm formation. Each LAB strain was tested on a separate plate in which four different groups (6 wells/group) were treated as follows:
-
- Test group: fill each well with 100 μl of S. mutans dilution, 100 μl of LAB dilution, and 800 μl of BHB media, subsequently.
-
- Control group I: fill each well with 100 μl of S. mutans dilution and 900 μl of BHB media, subsequently.
-
- Control group II: fill each well with 100 μl of LAB dilution and 900 μl of BHB media, subsequently.
-
- Blank group: fill each well with 1000 μl of BHB media.
After incubating these plates anaerobically at 37°C in 48 h, gently remove all the culture and residues from wells, then rinse the wells three times with 1000 μl of distilled water. The biofilms attached to wells were stained by adding 1000 μl of 0.1% crystal violet solution and incubating 30 minutes at room temperature. Gently remove all the stain solution and wash the wells again several times with 1000 μl of distilled water until OD590nm of the washed solution is lesser than 0.15. Finally, add to each well 1000 μl of 70% ethanol solution, mix gently and incubate 15 minutes at room temperature to completely dissolve the crystal violet incorporating in biofilm bacteria. Measure the optical density of ethanol solution from each well at 590 nm, then calculate the average OD590nm of each treated group. The ability of LAB strain to reduce S. mutans biofilm formation would be inferred if the average OD590 of test group and control group II were lower than that was from control group I (done by t-test for means, p < 0.05, Microsoft Excel 2016). The formula using to calculate the percentage of S. mutans biofilm reduction of certain LAB strain is described as below:
In which, OD590t, OD590c2 and OD590b were average OD590nm values of test group, control group II and blank group, respestively.
The resistance to artificial gastrointestinal conditions of LAB were assessed based on counting the viable LAB on MRS agar after short periods (1, 2, and 3 hours) of exposure to simulated solutions. For oral cavity condition, artificial saliva was made of 4000 UI/ml amylase in PBS solution. For mimicking gastric conditions, PBS solutions consisting of 3 mg/ml pepsin at pH 1.5 or 2.0 were used. For small instestine simulation, MRS broths supplemented with 0.5%, 1%, and 3% bile salt were used.
Minimal inhibitory concentration (MIC) of seven antibiotics including Penicillin, Ampicillin, Imipenem, Gentamycin, Vancomycin, Erythromycin, and Clindamycin on isolated LAB were detected using Macrodilution (tube) broth method following the instruction of M45 guideline (3rd edition), CLSI, 2016 [26]. Briefly, twofold dilution of antibiotic in Cation-adjusted Muller Hinton Broth (CAMHB) supplement with 5% lysed horse blood was prepared volumetrically in serial of 13 × 100 mm tubes to achieve final concentrations ranging from 128 to 0.125 μg/ml of each antimicrobial agent. The final volume of the dilution in test tubes was 1 ml. Transfer to each tube 1 ml of lactic acid bacteria inoculum prepared as described in section 2.1 and incubate these tubes anaerobically at 37°C in 24 h. Read the tubes in the dark background by unaided eyes and record the MIC as the lowest concentration of antimicrobial agent that completely inhibits the growth of bacteria in the tube.
LAB strains that showed the most potential in previous experiments were further examined the ability to reduce S.mutans biofilm formation by SEM. Human teeth were carefully scraped and cut into pieces of 0.5 cm × 0.5 cm and were used as scaffolds for biofilm formation of S. mutans. After 24 hours of incubating in the eppendorf containing S. mutans with or without the presence of tested LAB strains in BHB media (final density approx 107 CFU/ml for each bacteria), tooth pieces were gently rinsed with distilled water, then fixed with 2.5% glutaraldehyde and 4% paraformaldehyde, dehydrated in graded ethanol solutions (30 – 100%), and dried in ambient air. Finally, these tooth pieces were sent to SEM service at Nanotechnology Research Lab (Saigon Hi-tech Park, Ho Chi Minh City, VietNam). There, biofilm on tooth pieces would be sputter-coated with platin and observed using SEM at 3000× magnification. Tooth pieces without bacterial treatments were used as blank samples in this experiment.
Potential LAB strains were evaluated their effects on the expression of S. mutans forming genes including gtfB and luxS. The biofilm formation was carried out in a 50 ml Falcon tube using a glass slide as a scaffold and 40 ml of BHB as growth medium. In the test group, 500 μl of both LAB and S. mutans inoculum (each at density of approx. 108 CFU/ml, preprared as described in section 2.1) were added to the tube. In the control group, S. mutans inoculum was added solely. After 48 hours of incubation, biofilm attached in each glass slide was scratched off and resuspended in PBS. Bacteria were detached from biofilm by sonicating twice the biofilm suspension at energy level 3 for 25 seconds, with 2 minutes placing sample on ice between treatments. Then, biofilm samples were stored at -80°C in RNase free tubes until total RNA extraction. Next, total RNA were extracted from 250 μl of biofilm samples using 750 μl of TRIsureTM reagent (Bioline, UK) according to the the instruction of manufacturer and then, were assessed in 2% agarose gel electrophoresis as well as qualified using UV absorbance at 260 nm and 280 nm.
The expression level of gtfB, luxS, and ldh (internal control genes for qPCR normalizing) of S. mutans when co-culturing with LAB were analysed using two-step RT-qPCR. Firstly, cDNA of mRNA of these genes were synthesised from total RNA extract using ProtoScript® II First Strand cDNA Synthesis Kit (NEB, UK) following manufacturer protocol harboring specific primers (Table 2.). Next, qPCR reactions were performed using Exicycler™ 96 qPCR system (Bioneer, Korea). The amount of cDNA (gtfB, luxS, and ldh) was separately quantified in 25 μl reaction mixture containing 12.5 μl of SensiFASTTM SYBR® Hi-ROX Kit (Bioline, UK), 0.5 μl of each primer (50 μM), 1 μl of cDNA sample, 0.2 μl of GoTaq® Flexi DNA Polymerase (5 U/μl, Promega, USA) and 9.8 μl sterile deionized water. The thermal cycling program was set at follows: initial denaturation at 95°C for 2 mins followed by 40 cycles of 95°C for 30 secs, 55°C for 30 secs, and 72°C for 20 secs. Ct value of each gene was recorded and relative quantification was done by using the 2-ΔΔt method.
LAB dilution was prepared as described in section 2.1 but its OD600 was adjusted between 0.3 – 0.4 instead. Then, transfer the inoculum into each tube of API 50 CHL test strip and complete the test following the manufacturer’s protocol. The carbohydrate assimilation profile of each strain was recorded and analysed using ApiwebTM software.
LAB genome was extracted using Wizard® SV Genomic DNA purification system (Promega, USA) following the manufacturer’s manual. 16S rDNA gene was amplified from genome extract using pair of universal primers 27F (5’- AGAGTTTGATCCTGGCTC-3’) and 1492R (5’-TACGGTTACCTTGTTACGACT-3’). PCR reactions were conducted in 200 μl tube consisted of 5.0 μl of 10X Colorless GoTaq® Flexi Buffer (Promega, USA), 1.0 μl of 10 mM dNTPs mixtures, 0.5 μl of each primer (100 μM), 0.2 μl of GoTaq® Flexi DNA Polymerase (5 U/μl, Promega, USA) and 1 μl of genome template. The thermal cycling program was set for 30 cycles as follows: initial denaturation at 95°C for 5 mins, denaturation at 95°C for 45 secs, annealing at 55°C for 45 secs, elongation at 72°C for 2 mins and final elongation at 72°C in 10 mins. The PCR products were checked on 1% agarose electrophoresis and purified using Column PureTM PCR Clean-up Kit (ABM, USA) according to the manufacturer’s manual. Then, purified PCR products were sent to sequencing service at PHUSA Biochem (Can Tho city, VietNam) using pair of primers 27F and 1492R. Raw sequence of segments received were assembled using Seqman v7.1.0 software (Lasergene, DNASTAR, USA). Final assemblies were searched for similarity in NCBI data using BLAST tool.
3. RESULTS
From fermented vegetables, 51 isolates showing clear zones around colonies in MRS + CaCO3 agar were collected and isolated subsequently on MRS agar. Next, 36 of 51 isolates exhibiting basic properties of Lactobacilli (i.e. macroscopic and microscopic morphology, basic biochemical tests) were preliminary identified as Lactobacillus spp. strains. Similarly, the other 15 Lactobacillus spp. strains were isolated from healthy human saliva samples. In summary, 51 strains of Lactobacilli were successfully isolated and stored at -80°C.
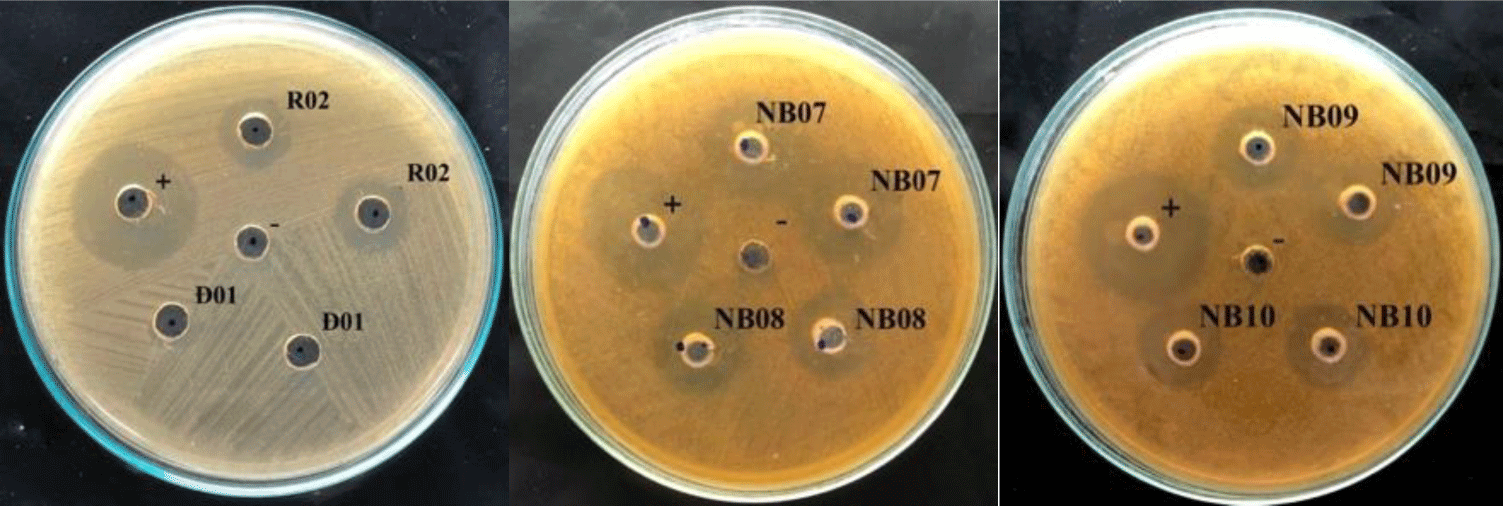
In the agar well diffusion assay, 14 out of 51 isolates showed the inhibiting effect on S. mutans growth, with inhibitory zones ranging from 8 – 12 mm, therein 7 strains were isolated from fermented vegetables and the others were from saliva. From that, 14 strains, includes K02, K03, C04, C05, R02, DH03, NB02, NB03, NB08, NB09, NB10 and NB15 were selected for testing the ability in reducing biofilm formation of S. mutans.
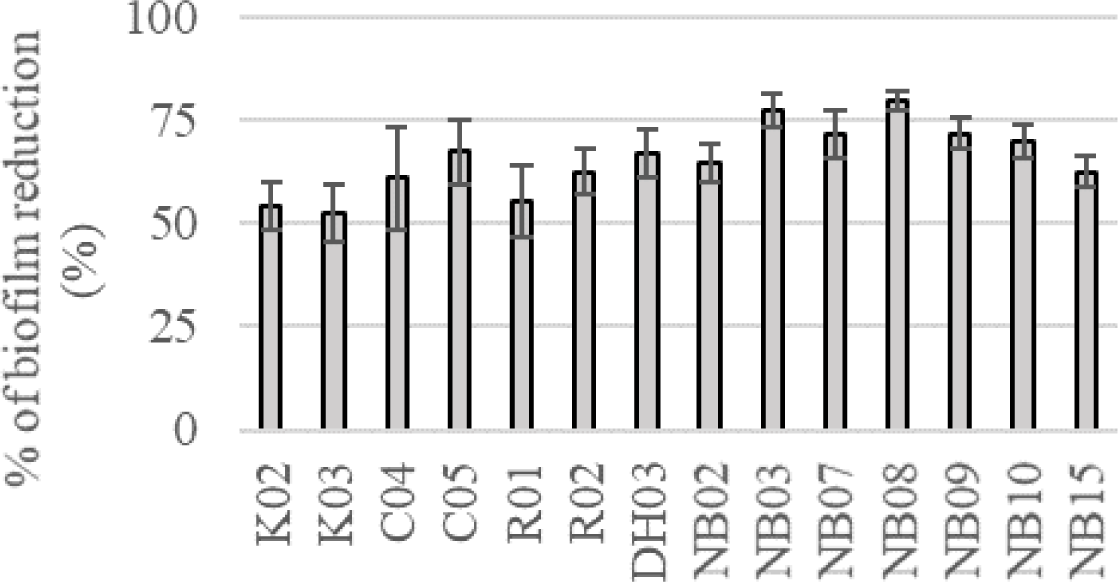
In the colorimetric assay, all of 14 tested strains could significantly reduce S. mutans biofilm formation by more than 50%, p < 0.05 (Figure 2). Especially, saliva originated strains (i.e. NB03, NB07, NB08, NB09, and NB10) could decrease the biofilm-forming level of S. mutans by more than 70%. These all 14 strains were used to evaluate probiotic properties.
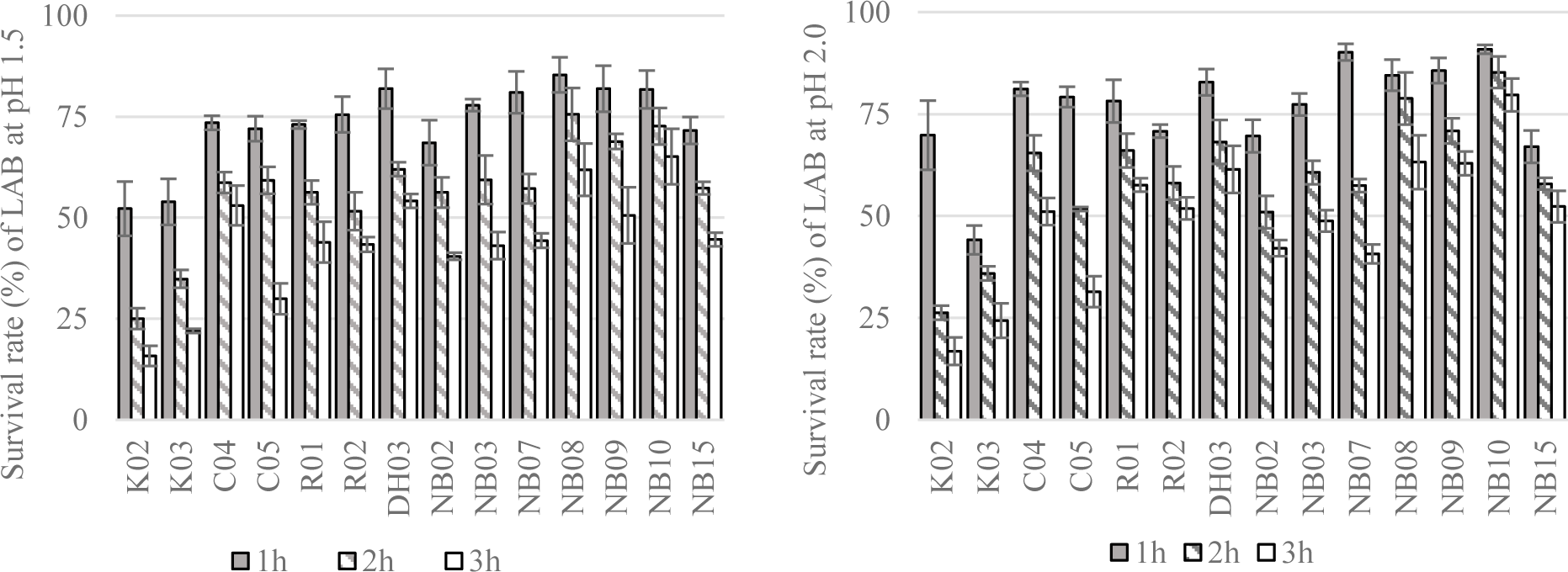
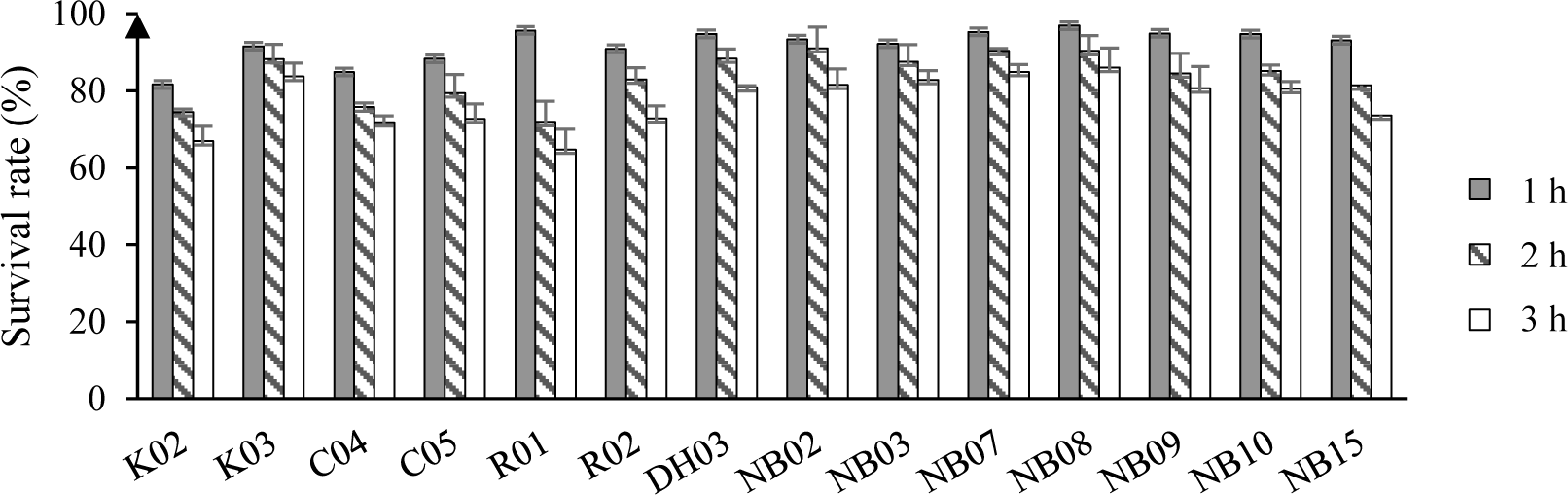
The survival rate of isolated LAB after a short time exposing to artificial saliva were displayed in Figure 3. Overall, the reduction in the number of bacteria were observed in all test strains. After 3 hour of exposure to 4000 IU/ml of amylase, there were 7 out of 14 strains showed survival rate above 80%, i.e. K03, DH03, NB02, NB07, NB09, and NB10. Most of the high survival rate strains were unsurprisingly isolated from human saliva.
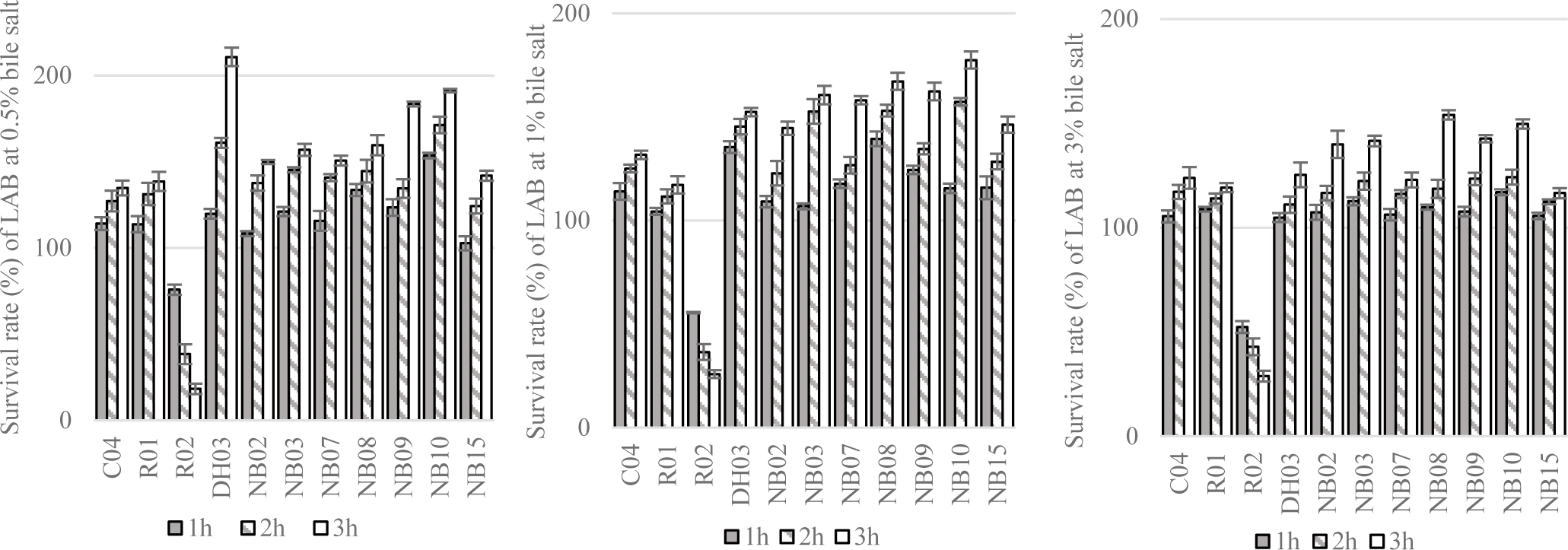
Next, the resistance of LAB to simulated gastric conditions at pH 1.5 or 2.0 were demonstrated in Figure 4. From the result of this experiment, 11 strains showing the greatest alive ratio (more than 50% and 40% of survival rate after 2 hours and 3 hours of exposure to low pH conditions, respectively) were chosen to test the tolerance to different concentrations of bile salt.
The tolerance of tested strains to different concentrations of bile salt were shown in Figure 5. In general, 10 out of 11 tested strains, i.e. C04, R01, DH03, NB02, NB03, NB07, NB08, NB09, NB10, NB15 could grow in 0.5 – 3.0% of bile salt condition. Thus, these strains were selected to evaluate antimicrobial susceptibility.
According to the interpreting criteria of M45 (3rd ed., CLSI, 2016), all of the studied strains were susceptible to tested antimicrobial agents. MIC values of each antibiotic to test strains were shown in Table 3.
In summary, through assessment of probiotic characteristics, four LAB strains including DH03, NB08, NB09 and NB10 that showed the greatest resistant ability to simulated gastrointestinal conditions as well as susceptible to antibiotics were chosen to further test for reduction effect on S. mutans biofilm formation by SEM and RT-qPCR analysis.
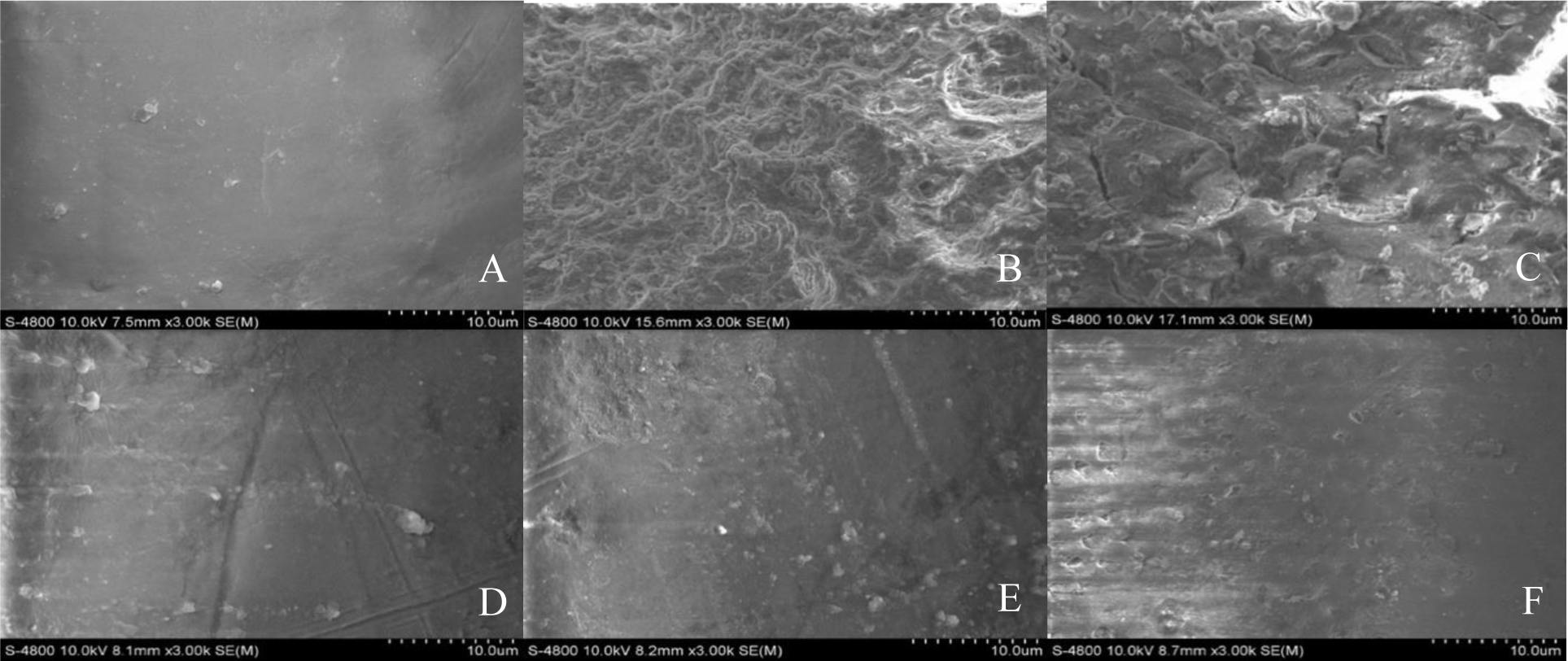
After inducing the formation of S. mutans biofilm on human teeth surface in the condition with and without the presence of tested LAB, biofilm structures were observed by SEM at 3000× magnification and showed in Figure 6. In the blank sample (Figure 6.A), the surface of tooth piece was smooth and no sign of bacterial growth and biofilm formation could be observed. Whereas, in the control sample, in which S. mutans was grown solely, biofilm was formed as microstructure like networks or dense slime (Figure 6.B). The amount of biofilm microstructure was significantly reduced in DH03 co-culturing sample (Figure 6.C), and almost eliminated when S. mutans was co-cultured with NB08, NB09 or NB10 strains (Figure 6.D, E, and F). Therefore, all of the tested strains exhibited the ability to reduce the biofilm formation of S. mutans.
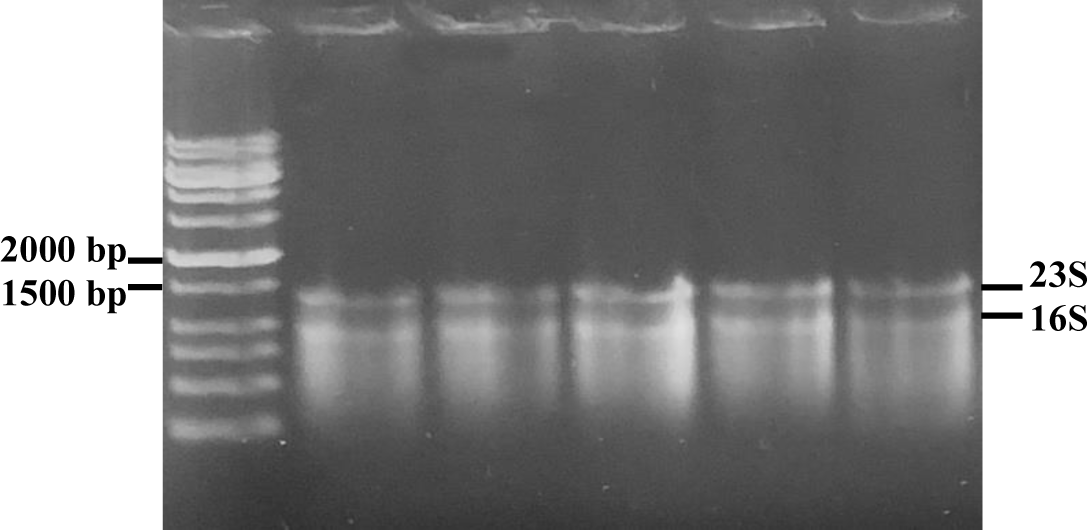
The analysis of total RNA extract from biofilm samples were shown in Figure 7. In all samples, the ratios of OD260/280 were greater than 2.0 and therefore RNA extracts were qualified to use in RT-qPCR reactions.
After conducting RT-qPCR to relative quantify of mRNA of target genes (gtfB, luxS) and normalizing gene (ldh), △△Ct of each gene were showed in Table 4. According to the result, DH03 could reduce the expression of both target S. mutans genes (81% and 87% of the expression level of gtfB and luxS compared to the untreated group, respectively). While, NB08 and NB09 were able to decrease the expression level of single gene to untreated samples (71% of the expression level of luxS and 79% of the expression level of gtfB in samples of coculturing S. mutans with NB08 and NB09, respectively). Co-culturing S. mutans with NB10 did not affect the expression of both target genes of S. mutans.
| Strain | Target genes | Expression level compared to untreatment (%) |
|---|---|---|
| DH03 | gtfB | 81% |
| luxS | 87% | |
| NB08 | gtfB | 102% |
| luxS | 71% | |
| NB09 | gtfB | 79% |
| luxS | 102% | |
| NB10 | gtfB | 94% |
| luxS | 98% |
The results of phenotypic and genotypic identification of the four lactic acid bacteria showed in Table 5.
According to carbohydrate assimilation profiles acquired from API 50CHL test strips, DH03, NB08, and NB10 were phenotypically identified as Lactobacillus spp. with %ID of 99.99%, 99.99% and 97.9%, respectively. NB09, however, appeared to be doubtfully identified as Lactobacillus brevis (with %ID of 71.5%) or Weissella confusa (with %ID of 28.3%). The reference strain Lactobacillus acidophilus ATCC 4356 was correctly identified to species level with %ID of 98.2%.
Partial 16S rDNA sequences of DH03, NB08, NB09, and NB10 were checked the similarity with DNA sequences in GeneBank by BLAST tool. The alignment results showed highly match to bank sequences with %ID of 99.05% to 100%.
4. DISCUSSION
With increasing demands for probiotic consuming, there were continuing efforts on searching and screening for new probiotic strains that showed potential effects on human and animal health in the last 10 years. Lactobacillus genus was among the most attractive probiotic microorganisms due to their proven health benefits and safety. Lactobacilli were long known for showing extensively antimicrobial activities on various pathogens by a number of mechanisms such as competing for colonization sites and nutrients; inhibiting pathogens spread; or producing various types of secondary metabolites like organic acids, hydrogen peroxide, bacteriocin or biosurfactants… Besides conventional sources such as milk, dairy products, or gut of humans and animals, recent studies showed that fermented vegetables were also potential sources for isolating lactic acid bacteria. Tamang et al., (2005) were successful in isolating different species of LAB that exhibited probiotic properties such as L. brevis, L. plantarum, Pediococcus pentosaceus… from fermented vegetables [28]. Swain et al., (2014) concluded that traditional Asia fermented vegetables and fruits were rich in LAB such as L. plantarum,L. pentosus, L. brevis, L. fermentum and L. casei [29]. In addition, sites of the human body other than gut were also sources for searching for new probiotics. Various potential LAB were found in the oral cavity, especially in human saliva [30]. From the oral cavity, Ko ll et al., (2008) isolated different Lactobacilli strains that exhibited antimicrobial effect on various dental pathogens such as L. casei, L. plantarum, L. rhamnosus [31]. Kang et al., (2006) cultivated Weisella cibaria from children saliva that could reduce the production of S. mutans biofilm [32]. Bosch et al., (2012) also found many LAB strains from human saliva that showed beneficial activities on oral health [33]. From the above discussions, fermented vegetables and healthy human saliva were used as sources to isolate potential LAB that exhibited antagonistic activity against S. mutans in this study. Through set of experiments, 51 LAB strains were successfully isolated and screened for inhibiting S. mutans growth and biofilm formation. Using the agar well diffusion method, 14 isolates were detected to inhibit S. mutans growth. This finding was in agreement with other studies in the literature about antagonism effects of LAB to many pathogens including S. mutans [14-23].
In order to evaluate the influences of these LAB in biofilm production of S. mutans, three different methods were used. First, using the colorimetric assay, all of 14 strains showed antibiofilm activity on S. mutans. Because of simplicity, this method was adopted by several researchers for studying the effects of Lactobacilli on S. mutans biofilm formation and also produced similar results [34, 35]. Among these strains, DH03, NB08, NB09, and NB10 showed their best in serial of probiotic characteristics evaluation tests, were chosen to further confirm the antibiofilm effect by SEM observation and RT-qPCR analysis. While SEM observation would give the exact quantitative information about biofilm formation, RT-qPCR would give more insight from molecular interactions between microbes. After conducting biofilm formation in the dual-species growth model, S. mutans biofilm was found to be decreased in the formation when co-culturing with DH03 and eliminated when co-culturing with either NB08, NB09, or NB10. By using SEM to study the influence of Lactobacillus spp. on S. mutans biofilm formation in various types of surface, similar observations were obtained from R. Wasfi et al., (2018) using glass slide as substratum [35] or Rossoni et al., (2018) using hydroxyapatite disks as substratum [36]. In RT-qPCR analysis of gtfB and luxS - crucial genes involved in biofilm production of S. mutans, DH03 was the only strain that could down-regulate the expression level of both target genes; while NB08 and NB09 affected just one or the other gene. NB10 showed no effect on the expression of both gtfB and luxS. Thus, the ability of Lactobacilli in reducing biofilm formation by regulating S. mutans genes depends on specific features of each strain. Gene ldh was used to normalize qPCR results in this study since it plays a crucial role in the survival of S. mutans and also its expression level is not affected by culture conditions [37]. The ability of Lactobacilli on alteration of the expression of S. mutans genes also reported by some group of researchers. A. Ahmed et al., (2014) reported that L. acidophilus strains could down-regulate both gtfB and luxS expression by 60-80% [34]. R. Wasfi et al., (2018) evaluated the effect of Lactobacillus spp. on different groups of biofilm regulator genes include EPS production genes (gtfB, gtfC, gtfD, and sacB), quorum-sensing modulating genes (comC, comD, vicK, vicR) and stress survival associating genes (atpD and aguD). The results showed that the expression levels of these biofilm assiociating genes were affected by not only the nature of Lactobacilli strains but also the state of S. mutans (planktonic or biofilm bacteria) [35]. Therefore, more studies must be conducted in order to obtain more insight into the interaction of the four LAB strains in this study and gene expression trait of S. mutans.
The species identification by phenotypic and genotypic methods of DH03, NB08, and NB10 produced similar results. NB09, however, was got unsatisfactory matching results between phenotypic and genotypic identification. The occasional difference in identification result between both methods was found in many studies of Lactobacilli identification. E. M. Brolazo et al., (2011) found that Lactobacillus spp. identification by conventional biochemical tests was not matching with the genotypic method and suggested that the genotypic method was more reliable with lower failure rate than the biochemical method [38]. P. Moraes et al., (2013) compared the results of LAB identification by 16S rDNA sequencing to API 50 CHL kits and concluded that genotypic identification was more accurate and reliable. The lacking of reliability of API 50 CHL system is due the manufacturer’s database is not frequently updated and some Lactobacillus species are missing. From the above arguments, the four tested strains were assigned by genotypic method as L. plantarum (DH03 and NB08), L. brevis (NB10), and W. confusa (NB09).
L. plantarum showed potential characteristics in inhibiting growth and biofilm formation of S. mutans in many studies. According to L. Xiaolong et al., (2017), L. plantarum ST-III could inhibit S. mutans growth and reduce biofilm formation up to 80% [41]. There were different mechanisms suggested for the antagonism effects of L. plantarum on S. mutans [18-20]. Moreover, L. plantarum also showed many advantages as probiotic such as antimicrobial effects on various pathogens, high survival rate in gut conditions [42, 43].
L. brevis strains were also reported in some studies about inhibitory effects on different Streptococci includes S. mutans. Fang et al., (2018) reported that L. brevis could produce sorts of secondary metabolites to suppress S. mutans growth [43]. L. brevis also features useful properties that is suitable for use as oral probiotic such as the ability to adhere to oral epithelial cells, anti-inflammation effects, and immune-modulating [43, 44].
W. confusa (basonym: Lactobacillus confusus) belongs to the genus Weissella, Leuconostocaceae family, and is considered as lactic acid bacteria [45]. W. confusa could be isolated from human sources (breast milk, feces, saliva…) or different types of fermented foods [46 - 48]. Studies showed that Weissella species have closely phylogenetic relationships with Lactobacillus and Leuconostoc species and it is challenging in differentiating these species by phenotypic characteristics. Thus, genetic analysis approach must be used to identify these bacteria [49]. W. confusa strains are also considered as potential probiotic. W. confusa was reported to reduce biofilm formation of pathogens up to 90% and also showed great tolerance in acidic or bile salt conditions [48]. Moreover, Weissella species isolated from saliva also suppressed more than 90% of biofilm production in S. mutans and were predicted to exhibit physiological pH that was more suitable to oral cavity than other Lactobacilli [32]. In safety concerns, there were some case reports about opportunistic infection caused by W. confusa in immunodeficient patients [50, 51]. However, to those bacteria classified as GRAS such as L. casei or L. rhamnosus, there were also reports about opportunistic infection caused by them [52]. Therefore, the safeness of each lactic acid bacteria in general and W. confusa in particular must be evaluated carefully in in vivo models.
Besides the above experiments, we also tested the ability to metabolize sucrose of the four strains DH03, NB08, NB09, and NB10. The result showed that W. confusa NB09 and L. brevis NB10 were sucrose negative strains (data not shown). Tooth decay and the growth of cariogenic bacteria like S. mutans were strictly related to carbohydrate-rich diets, especially sucrose – the most common sugar used today. Therefore, the selection of probiotic strains that showed not only antagonistic effects on S. mutans but also did not ferment sucrose could be an important advantage in reducing the acid load in sweet tooth persons.
5. CONCLUSION
In this current study, we were successful in isolation of lactic acid bacteria, i.e. L. plantarum DH03, L. plantarum NB08, W. confusa NB09 and L. brevis NB10, that showed inhibitory effects on S. mutans growth as well as reducing biofilm formation. These four strains also showed appropriate characteristics of probiotics such as high tolerance to artificial gastrointestinal conditions and susceptible to antibiotics. The result of this study will be a basis for preparation of lozenges or chewing gum containing these strains in order to prevent dental plaque initiated by S. mutans. However, further studies must be conducted in order to evaluate their efficiency and safeness in in vivo models.